Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 10(1); 2019 > Article
-
Original Article
Specification of Bacteriophage Isolated Against Clinical Methicillin-ResistantStaphylococcus Aureus - Ahmad Nassera,b, Reza Aziziana, Mohsen Tabasic, Jamil Kheirvari Khezerlood, Fatemah Sadeghpour Heravie, Morovat Taheri Kalania, Norkhoda Sadeghifarda, Razieh Aminif, Iraj Pakzada, Amin Radmaneshc, Farid Azizi Jaliliang
-
Osong Public Health and Research Perspectives 2019;10(1):20-24.
DOI: https://doi.org/10.24171/j.phrp.2019.10.1.05
Published online: February 28, 2019
aClinical Microbiology Research Center, Ilam University of Medical Sciences, Ilam, Iran
bDepartment of Pathobiology, School of Public Health, Tehran University of Medical Sciences, Poursina St., Tehran, Iran
cLegal Medicine Research Center, Legal Medicine Organization, Tehran, Iran
dYoung Researchers and Elite Club, Tehran Medical Sciences, Islamic Azad University, Tehran, Iran
ePertussis Reference Laboratory‚ Department of Bacteriology, Pasteur Institute of Iran, Tehran, Iran
fDepartment of Molecular Medicine, School of Medicine, Hamadan University of Medical Sciences, Hamadan, Iran
gDepartment of Virology, School of Medicine, Hamadan University of Medical Sciences, Hamadan, Iran
- *Corresponding author: Farid Azizi Jalilian, Department of Virology, School of Medicine, Hamadan University of Medical Sciences, Hamadan, Iran, E-mail: azizijalilian@yahoo.com, azizifarid@gmail.com
Copyright ©2019, Korea Centers for Disease Control and Prevention
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Abstract
-
Objectives
- The emergence of resistant bacteria is being increasingly reported around the world, potentially threatening millions of lives. Amongst resistant bacteria, methicillin-resistant Staphylococcus aureus (MRSA) is the most challenging to treat. This is due to emergent MRSA strains and less effective traditional antibiotic therapies to Staphylococcal infections. The use of bacteriophages (phages) against MRSA is a new, potential alternate therapy. In this study, morphology, genetic and protein structure of lytic phages against MRSA have been analysed.
-
Methods
- Isolation of livestock and sewage bacteriophages were performed using 0.4 μm membrane filters. Plaque assays were used to determine phage quantification by double layer agar method. Pure plaques were then amplified for further characterization. Sulfate-polyacrylamide gel electrophoresis and random amplification of polymorphic DNA were run for protein evaluation, and genotyping respectively. Transmission electron microscope was also used to detect the structure and taxonomic classification of phage visually.
-
Results
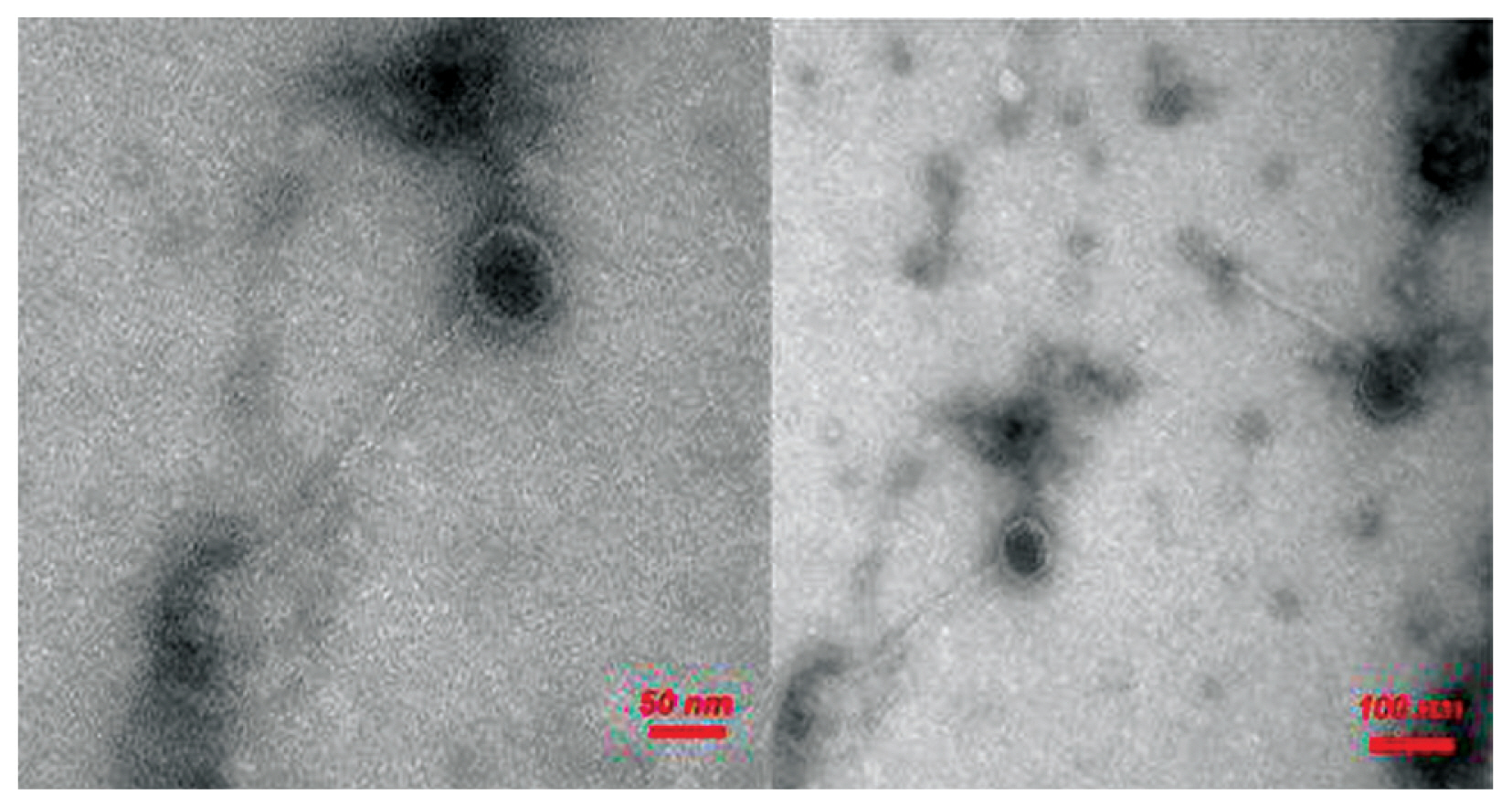
- Head and tail morphology of bacteriophages against MRSA were identified by transmission electron microscopy and assigned to the Siphoviridae family and the Caudovirales order.
-
Conclusion
- Bacteriophages are the most abundant microorganism on Earth and coexist with the bacterial population. They can destroy bacterial cells successfully and effectively. They cannot enter mammalian cells which saves the eukaryotic cells from lytic phage activity. In conclusion, phage therapy may have many potential applications in microbiology and human medicine with no side effect on eukaryotic cells.
- Staphylococcus aureus is a gram-positive, non-motile, non-sporulating bacterium which causes hospital and community infections [1]. This bacterium is responsible for several life threatening infections including endocarditis, osteomyelitis, skin and soft tissue infections and wound infections leading to a high mortality rate each year [2]. It has been reported that mortality of methicillin-resistant Staphylococcus aureus (MRSA) infections have increased amongst AIDS patients in the United States of America [3]. Based on a systematic review and meta-analysis in Iran, 7,464 samples of S. aureus were isolated from patients from different cities, with 52.7% ± 4.7% of strains carried the mecA gene which is responsible for resistance to methicillin [4].
- Staphylococcus can induce cellular apoptosis after invasion of host cells, and invasion mediated by Staphylococcal fibronectin-binding proteins allow the other microorganisms to bind to the hosts cells and initiate secondary infections [5]. Due to the methicillin resistance of S. aureus to most commonly used antibiotics such as oxacillin, nafcillin, methicillin and even vancomycin, treatment of MRSA is more challenging compared to other resistant strains [6]. In addition, indiscriminate and overuse of antibiotics has made the penicillin susceptible S. aureus strains, resistant to the population to ß-lactam antibiotics [7].
- Bacteriophages (phages) can be found in all habitats and more significantly colonization of bacterial strains. A phage is a type of virus that can attack and destroy bacterial cells [8]. Twelve distinct groups of phages have been discovered which are highly specific against bacterial species [9]. The order of Caudovirales bacteriophages can be categorized into 3 major families including Siphoviridae, Myoviridae, and Podoviridae. All members of the Caudovirales order, have double stranded DNA, with a morphological head and tail structure [9]. The typical structure of a phage includes a head that is filled with DNA or RNA, and a tail used for injection of the genome into the bacterial cells. Due to receptor limitations on phages, they are unable to infect mammalian cells [10].
- Isolation of phages against many bacterial populations have been carried out from wastewater, sewage, retail raw chicken meat, soil, blood cultures, cerebrospinal fluid, superficial wounds, deep wound swabs, respiratory tract specimens, urine and miscellaneous specimens [8]. After the process of phage isolation specifically against MRSA, they can be used as an alternative therapy to treat MRSA infections. Due to the inability of bacteriophages to enter mammalian cells, no side effects on eukaryotic cells have been reported [11].
- The main aim of this study was to isolate and characterize lytic phages against MRSA reporting their morphology, genetic material and structural protein composition.
Introduction
- 1. Bacterial strain and growth media
- Bacterial strains were isolated from clinical specimens from hospitals in the west of Iran, Kermanshah. Isolates were incubated at 37°C for 24 hours on blood agar (Oxoid Ltd, Basingstoke, UK) and then sub-cultured on TS agar (Tryptic Soya, Liofil-chem, Italy). Single colonies were tested with tube coagulase and the DNase test, and growth on mannitol salt agar (Difco, Detroit, Mich., USA).
- After identification of S. aureus positive specimens, they were stored in 20% glycerol at −20°C. The Oxacillin susceptibility test was performed to identify MRSA strains (0.5 McFarland suspension of S. aureus was spotted onto Mueller-Hinton agar supplemented with 4% NaCl and 6 ug of oxacillin per mL). To initiate the analysis, subculture bacterial strains were added to the nutrient agar. In addition, the presence of the methicillin resistance gene (mecA) was determined by multiplex polymerase chain reaction (PCR; Eppendorf epGradient S thermocycler).
- 2. Isolation of phages against MRSA
- Phages against clinical MRSA in sewage and stool samples were isolated based on the enrichment method described by Cerveny, et al with some modifications [12]. Briefly, sewage and stool samples were collected from the main sewage pipe and livestock farming in Ilam University of Medical Science and Ilam University, respectively. Samples were centrifuged and supernatants were filtered through 0.45 and 0.2 μm sterile filters. Twenty mL of filtered sewage sample and 20 mL sterile nutrient broth were mixed and spiked with 5.0 mL of overnight culture clinical MRSA, and then incubated at 37°C for 24 hours. To deplete bacterial strains, the mixture had undergone centrifugation and the supernatant was then sterilized by 0.45 and 0.2 μm filters. Moreover, to check for the presence of phages, a double layer agar method was used as described by Adams [13].
- Double agar overlay technique was performed to determine plaque formation. Each of the phage suspensions was serially diluted. This was performed by adding 1 mL of diluted phage and 1 mL of host bacterium that was mixed with 2 mL of molten soft-agar (0.75 % agar, w/v). This was poured quickly onto the surface of a solidified nutrient agar plate (1/5 % agar, w/v; Difco Laboratories) [13].
- All of the isolated phages were purified by single-plaque isolation, and mix plaques with CaCl2 were obtained by the standard procedure presented by Sambrook et al [14]. One separated phage was picked with a sterile pipette tip along with the surrounding cell mass, and then inoculated into100 μL CaCl2 with 200 μL of host strain that had been cultured overnight. One mL of top agar was added and quickly poured onto a solidified nutrient agar plate.
- Phages were concentrated based on the published methods described by Yamamoto et al [15] with some modifications. MRSA isolated from host cells was added to the isolated phages and then incubated for 24–48 hours at 37°C to complete the lysis of host cells. In the next step, culture fluid was centrifuged and filtered through 0.4 μm filters. Polyethylene glycol 8,000 was added to make the final concentration of 20%. Finally, the pellet was collected following centrifugation (12,000 rpm for 20 minutes in 4°C).
- 3. Random amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR) analysis
- Phage DNA was extracted by DNA extraction kit (NOREGEN®). RAPD-PCR was performed based on the Johansson et al [16] study with some modifications.
- Four different primers used in this study which included: primer 1: (5′-ACGCAGGCAC-3′), primer 2: (5′-AACGCGCAAC-3′), primer 3: (5′-CCGCAGCCAA-3′) and primer 4: (5′-AACGGGCAGA-3′) that were selected based on a relatively similar study [17]. RAPD-PCR cycles are described in Table 1.
- The PCR mixture consisted of 1 μL Taq DNA polymerase, 1 μL primer, 1 μL dNTPs, 2 μL PCR buffer, 1 μL phage DNA, and 6.5 μL distilled water (for 12.5 μL reaction). DNA band patterns of each primer were obtained after gel electrophoresis (0.8% agarose gel) of the RAPD-PCR (12.5 μL). DNA molecular weight marker (1,000 bp) was used as a standard. Four types of MRSA phage were isolated from the stool samples.
- 4. Sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)
- Phage sedimentation (with 20% PEG and 12,000 rpm, 20 minutes in 4°C) was used for sodium dodecyl SDS-PAGE separation. Formaldehyde was used to prevent the effects of PEG on SDS-PAGE.
- MRSA-specific phages were mixed with an equal volume of sample buffer (0.0625 M Tris-HCl; pH 6.8, 15% glycerol, 1% SDS, 1% Beta-mercaptoethanol and bromophenol blue), and heated in a boiling water bath for 10–15 minutes [18].
- 5. Transmission Electron Microscopy
- After transferring the samples containing CaCl2 to the laboratory, samples were prepared with uranyl acetate (2%) for transmission electron microscopy (TEM) performed at the TEM Lab, Institute of Biochemistry and Biophysics, University of Tehran, Tehran, Iran. Firstly, the same amount of sample and glutaraldehyde (2.5%) was placed on a carbonic grid. After 1.5 minutes, the grid was left to dry on filter paper then washed with deionized water. One drop of uranyl acetate (2%) was added to the sample, and the grid was dried out on filter papers; samples were then ready for TEM. Particles were examined using a ZEISS EM 900 electron microscope (80 kV accelerating voltage; TEM Lab, Nano Kefa Institute, Iran).
- 6. Host range
- The host range of each phage was determined by inoculation of bacterial species into a double-layer agar, and the phages were tested against all the clinical MRSA.
Materials and Methods
2.1. Plaque assay
2.2. Phage purification and concentration
- 1. Detection of mecA gene in clinical MRSA
- To determine the presence of the methicillin resistance gene (mecA) in clinical MRSA samples, multiplex PCR was used (Figure 1).
- 2. RAPD-PCR
- RAPD-PCR was performed on the phage-extracted DNA from stool samples. In this study, 4 different primers were used which differentiated between all MRSA specific phages (Figure 2).
- 3. Structural protein analysis
- The structure of MRSA specific phage proteins shown in Figure 3 revealed 2 major protein bands approximately 30 and 48 kDa in weight, and a minor structure protein band, of approximately 80 kDa (Figure 3).
- 4. Phage morphology
- Based on the morphological features of the bacteriophages observed by electron microscopy, all the phages examined showed a long icosahedral capsid and non-contractile tails. According to the International Committee on Taxonomy of viruses, these phages were belonged to the Siphoviride family [19] of the Caudovirales order [20]. The morphology of MRSA-specific bacteriophage has been illustrated in Figure 4.
Results
- S. aureus is a source of many human and animal infections such as osteomyelitis, endocarditis, skin abscesses and many more [2]. Overuse of antibiotics has led to the emergence of MRSA. The emergence of MRSA strains highlights the necessity for therapeutic improvements in the future [21]. It has been suggested that the combination of bacteriophages and antibiotic therapy maybe more effective than unilateral treatment [21]. Due to the failure of treatment of Staphylococcal infections by routinely used antibiotics, this study highlights the role of phage therapy as a possible solution [22]. Phage therapy has many potential applications in human medicine and dentistry as well as in veterinary science and agriculture, and it can be an appropriate alternative therapy especially for multi drug resistant bacteria [23, 24].
- The first report of phage therapy in clinical medicine was described by Felix d’Herelle [25]. Many researchers think bacteriophages can play an important role to defeat resistant bacteria such as S. aureus. There has been a remarkable increase in the number of Staphylococcus phages with marvellous progress in targeting resistant S. aureus [22].
- In a similar study, different MRSA-specific phages belonging to the Podoviridae family, have been isolated from farm animals, and visualized using electron microscopy which showed the possibility of using lytic bacteriophages against MRSA strains [26]. In another study, 2 new phages isolated from a farmyard were classified into the Siphoviridae family which had lytic activity against MRSA [27].
- There are a wide variety of phages on the planet capable in destroying a large number of different bacterial populations [23]. Compared to the classic antibiotics, phage therapy is more cost-effective and without serious side effects on eukaryotic cells. Phages have complementary receptors on bacterial cell membranes [28]. The specific affinity of bacteriophages to the particular bacterial strains can prevent common side effects reported from routine antibiotics therapy. In addition, phage gene modification can improve the specificity and sensitivity of bacteriophages against bacterial strains [29]. Phenotype variation can help bacteria to escape from immune system and anti-bacterial treatments [29]. This problem can also be addressed by using phage mutants [30].
- In this study, isolation and characterization of effective bacteriophages with lytic activity against MRSA has been evaluated. Based on our finding, phage therapy could be a good candidate to combine with traditional therapies.
- However, to gain the benefits from the natural action of bacteriophages against the bacterial population, more clinical and in vivo studies should be conducted to show efficacy of phage therapy.
- In conclusion, the phages reported in this study exhibited good bacterial lysis activity. However, these isolated phages need to be further characterized and if found to have efficacy, they can be used in commercial lysate preparations in the near future, where therapeutic potential against a wider range of bacterial strains can be further investigated. Research using animal models of phage-bacterial interactions and ultimately clinical trials will highlight this topic and may provide a powerful alternative treatment against the dangers of multi-resistant pathogens.
Discussion
-
Acknowledgements
- This study has been supported by a research grant from Ilam University of Medical Sciences, Ilam, Iran.
Acknowledgments
- 1. Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev 2000;13(1). 16−34. PMID: 10.1128/CMR.13.1.16. PMID: 10627489. PMID: 88931.ArticlePubMedPMC
- 2. Lowy FD. Staphylococcus aureus infections. N Engl J Med 1998;339(8). 520−32. PMID: 10.1056/NEJM199808203390806. PMID: 9709046.ArticlePubMed
- 3. Stein R. [Internet]. Drug-resistant staph germ’s toll is higher than thought. Washington Post 2007 Available from: http://www.washingtonpost.com/wp-dyn/content/article/2007/10/16/AR2007101601392.html.
- 4. Askari E, Soleymani F, Arianpoor A, Tabatabai SM. Epidemiology of mecA-methicillin resistant Staphylococcus aureus (MRSA) in Iran: a systematic review and meta-analysis. Iran J Basic Med Sci 2012;15(5). 1010−9.PubMedPMC
- 5. Menzies BE. The role of fibronectin binding proteins in the pathogenesis of Staphylococcus aureus infections. Curr Opin Infect Dis 2003;16(3). 225−9. PMID: 10.1097/00001432-200306000-00007. PMID: 12821812.ArticlePubMed
- 6. Chambers HF, DeLeo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 2009;7(9). 629−41. PMID: 10.1038/nrmicro2200. PMID: 19680247. PMID: 2871281.ArticlePubMedPMCPDF
- 7. Enright M, DAR , Randle G, Feil E. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). PNAS 2002;7687−92. PMID: 10.1073/pnas.122108599. PMID: 12032344. PMID: 124322.ArticlePubMedPMC
- 8. Azizian R, Azizi Jalilian F, Sekawi Z, Amini R. Dynamics of Bacteriophages as a Promising Antibiofilm Agents. J Pure Appl Microbiol 2014;8(2). 1015−9.
- 9. Rezaei F, Nasser A, Azizi Jalilian F, Hobbs Z. Using Phage as A Highly Specific Antibiotic Alternative Against Methicillin Resistance Staphylococcus aureus (MRSA). Biosci Biotechnol Res Asia 2014;11(2). 523−9. PMID: 10.13005/bbra/1302.Article
- 10. Van Belleghem J, Dąbrowska K, Vaneechoutte M, Barr J, Bollyky P. Interactions between Bacteriophage, Bacteria, and the Mammalian Immune System. Viruses 2019;11(1). 1−22.Article
- 11. Miernikiewicz P, Dąbrowska K, Piotrowicz A, et al. T4 Phage and Its Head Surface Proteins Do Not Stimulate Inflammatory Mediator Production. PLoS ONE 2013;8(8). e71036PMID: 10.1371/journal.pone.0071036. PMID: 23976975. PMID: 3745418.ArticlePubMedPMC
- 12. Cerveny KE, DePaola A, Duckworth DH, Gulig PA. Phage therapy of local and systemic disease caused by Vibrio vulnificus in iron-dextran-treated mic. Infect Immun 2002;70(11). 6251−62. PMID: 10.1128/IAI.70.11.6251-6262.2002. PMID: 12379704. PMID: 130292.ArticlePubMedPMC
- 13. Gudina I, Gizachew Z, Woyessa D, Tefera TK. Isolation of Bacteriophage and Assessment of its Activity against Biofilms of Uropathogenic Escherichia coli in Jimma Town, South Western Ethiopia. Am J Curr Microbiol 2018;6(1). 52−66.
- 14. Petrenko V. Landscape phage: evolution from phage display to nanobiotechnology. Viruses 2018;10(6). 311PMID: 10.3390/v10060311. PMID: 6024655.ArticlePubMedPMC
- 15. Yamamoto KR, Alberts BM, Benzinger R, Lawhorne L. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virol 1970;40(3). 734−44. PMID: 10.1016/0042-6822(70)90218-7.Article
- 16. Johansson ML, Quednau M, Molin G, Ahrne S. Randomly amplified polymorphic DNA (RAPD) for rapid typing of Lactobacillus plantarum strains. J Appl Microbiol 1995;21(3). 155−9. PMID: 10.1111/j.1472-765X.1995.tb01030.x.Article
- 17. Gutierrez D, Martin-Platero AM, Rodriguez A, et al. Typing of bacteriophages by randomly amplified polymorphic DNA (RAPD)-PCR to assess genetic diversity. FEMS Microbiol Lett 2011;322(1). 90−7. PMID: 10.1111/j.1574-6968.2011.02342.x. PMID: 21692832.ArticlePubMedPDF
- 18. Kwiatek M, Parasion S, Mizak L, et al. Characterization of a bacteriophage, isolated from a cow with mastitis, that is lytic against Staphylococcus aureus strains. Arch Virol 2012;157(2). 225−34. PMID: 10.1007/s00705-011-1160-3.ArticlePubMedPDF
- 19. Maleki F, Hadadi MH, Rezaei F, et al. Classification and Replication Mechanism of Staphylococcus Phage. Biosci Biotechnol Res Asia 2015;12(1). 481−6. PMID: 10.13005/bbra/1689.Article
- 20. Ackermann H. Tailed bacteriophage: The order caudovirales. Adv Virus Res 1998;51:135−201. PMID: 10.1016/S0065-3527(08)60785-X.ArticlePubMedPMC
- 21. Wang Z, Zheng P, Ji W, et al. SLPW: A virulent bacteriophage targeting methicillin-resistant Staphylococcus aureus in vitro and in vivo. Front Microbiol 2016;7:934PMID: 27379064. PMID: 4908117.ArticlePubMedPMC
- 22. Nobrega FL, Costa AR, Kluskens LD, Azeredo J. Revisiting phage therapy: new applications for old resources. Trends Microbiol 2015;23(4). 185−91. PMID: 10.1016/j.tim.2015.01.006. PMID: 25708933.ArticlePubMed
- 23. Golkar Z, Bagasra O, Pace DG. Bacteriophage therapy: a potential solution for the antibiotic resistance crisis. J Infect Dev Ctries 2014;8(2). 129−36. PMID: 10.3855/jidc.3573. PMID: 24518621.ArticlePubMed
- 24. Rakhuba D, Kolomiets E, Dey ES, Novik G. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol 2010;59(3). 145−55. PMID: 21033576.ArticlePubMedPDF
- 25. Fruciano DE, Bourne S. Phage as an antimicrobial agent: d’Herelle’s heretical theories and their role in the decline of phage prophylaxis in the West. Can J Infect Dis Med Microbiol 2007;18(1). 19−26. PMID: 10.1155/2007/976850.ArticlePubMedPMCPDF
- 26. Kraushaar B, Thanh MD, Hammerl JA, et al. Isolation and characterization of phages with lytic activity against methicillin-resistant Staphylococcus aureus strains belonging to clonal complex 398. Arch Virol 2013;158(11). 2341−50. PMID: 10.1007/s00705-013-1707-6. PMID: 23760627.ArticlePubMedPDF
- 27. Vandamme EJ, Mortelmans K. A century of bacteriophage research and applications: impacts on biotechnology, health, ecology and the economy. J Chem Technol Biotechnol 2019;94(2). 323−42. PMID: 10.1002/jctb.5810.Article
- 28. Wang I-N, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol 2000;54:799−825. PMID: 10.1146/annurev.micro.54.1.799. PMID: 11018145.ArticlePubMed
- 29. Nobrega FL, Costa AR, Kluskens LD, Azeredo J. Revisiting phage therapy: new applications for old resources. Trends Microbiol 2015;23(4). 185−91. PMID: 10.1016/j.tim.2015.01.006. PMID: 25708933.ArticlePubMed
- 30. Samson JE, Magadán AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 2013;11(10). 675−87. PMID: 10.1038/nrmicro3096. PMID: 23979432.ArticlePubMedPDF
References
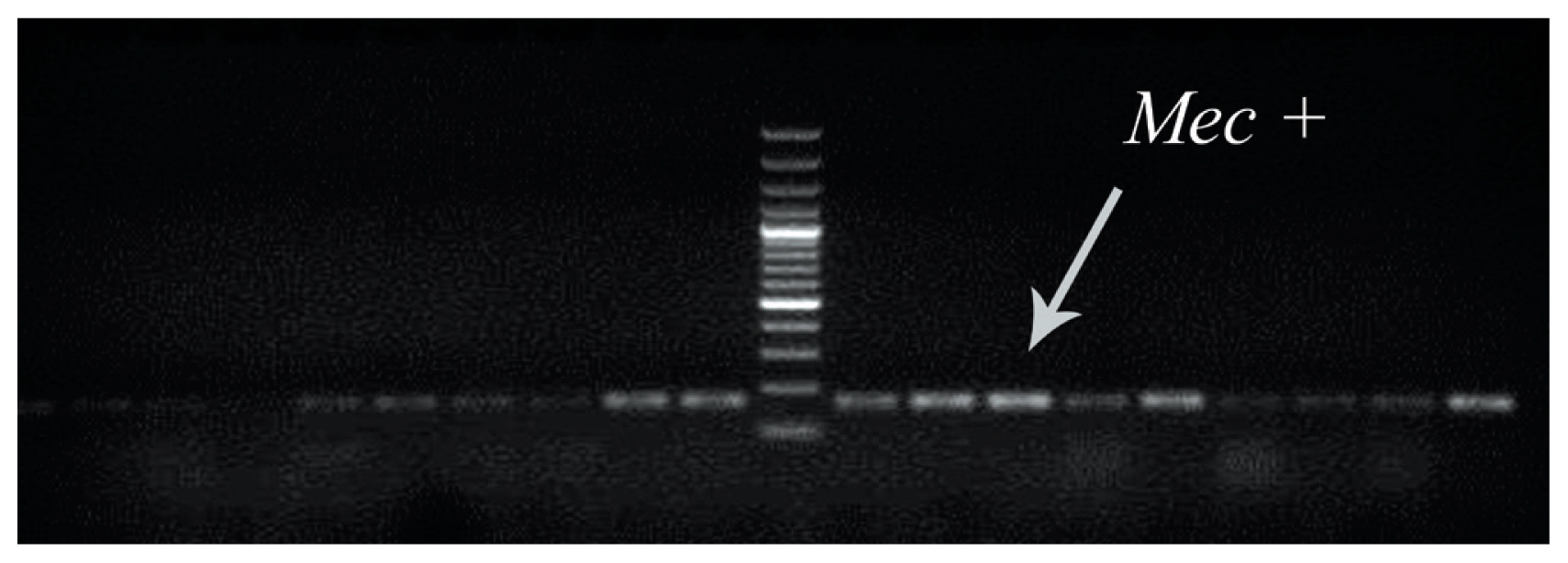


Figure & Data
References
Citations

- Characterization of a Vibrio parahaemolyticus-targeting lytic bacteriophage SSJ01 and its application in artificial seawater
Jungu Kang, Yoonjee Chang
Food Science and Biotechnology.2024; 33(6): 1505. CrossRef - Isolation and encapsulation of bacteriophage with chitosan nanoparticles for biocontrol of multidrug-resistant methicillin-resistant Staphylococcus aureus isolated from broiler poultry farms
Mona M. Elsayed, Rasha M. Elkenany, Ayman Y. EL-Khateeb, Nehal M. Nabil, Maram M. Tawakol, Heba M. Hassan
Scientific Reports.2024;[Epub] CrossRef - P1 Bacteriophage-Enabled Delivery of CRISPR-Cas9 Antimicrobial Activity Against Shigella flexneri
Yang W. Huan, Vincenzo Torraca, Russell Brown, Jidapha Fa-arun, Sydney L. Miles, Diego A. Oyarzún, Serge Mostowy, Baojun Wang
ACS Synthetic Biology.2023; 12(3): 709. CrossRef - Staphylococcus aureus Dormancy: Waiting for Insurgency
Ahmad Nasser, Shiva Jahanbakhshi, Mohammad Mehdi Soltan Dallal, Maryam Banar, Azin Sattari-Maraji, Taher Azimi
Current Pharmaceutical Biotechnology.2023; 24(15): 1898. CrossRef - Recent advance on nanoparticles or nanomaterials with anti-multidrug resistant bacteria and anti-bacterial biofilm properties: A systematic review
Farhad Moradi, Arshin Ghaedi, Zahra Fooladfar, Aida Bazrgar
Heliyon.2023; 9(11): e22105. CrossRef - Isolation and characterization of lytic bacteriophages from sewage at an egyptian tertiary care hospital against methicillin-resistant Staphylococcus aureus clinical isolates
Safia Samir, Amira El-Far, Hend Okasha, Rania Mahdy, Fatima Samir, Sami Nasr
Saudi Journal of Biological Sciences.2022; 29(5): 3097. CrossRef - Staphylococcus aureus: Biofilm Formation and Strategies Against it
Ahmad Nasser , Mohammad Mehdi Soltan Dallal, Shiva Jahanbakhshi, Taher Azimi, Leila Nikouei
Current Pharmaceutical Biotechnology.2022; 23(5): 664. CrossRef - An Anti-MRSA Phage From Raw Fish Rinse: Stability Evaluation and Production Optimization
Israa M. Abd-Allah, Ghadir S. El-Housseiny, Mohammad Y. Alshahrani, Samar S. El-Masry, Khaled M. Aboshanab, Nadia A. Hassouna
Frontiers in Cellular and Infection Microbiology.2022;[Epub] CrossRef - Molecular mechanisms of Shigella effector proteins: a common pathogen among diarrheic pediatric population
Ahmad Nasser, Mehrdad Mosadegh, Taher Azimi, Aref Shariati
Molecular and Cellular Pediatrics.2022;[Epub] CrossRef - A Metal-Containing NP Approach to Treat Methicillin-Resistant Staphylococcus aureus (MRSA): Prospects and Challenges
Wendy Wai Yeng Yeo, Sathiya Maran, Amanda Shen-Yee Kong, Wan-Hee Cheng, Swee-Hua Erin Lim, Jiun-Yan Loh, Kok-Song Lai
Materials.2022; 15(17): 5802. CrossRef - Electrochemical Biosensors for Pathogen Detection: An Updated Review
Morteza Banakar, Masoud Hamidi, Zohaib Khurshid, Muhammad Sohail Zafar, Janak Sapkota, Reza Azizian, Dinesh Rokaya
Biosensors.2022; 12(11): 927. CrossRef - Isolation of a lytic bacteriophage for Helicobacter pylori
Sara Khosravi, Razieh Amini, Mohammad Reza Arabestani, Seyed Saman Talebi, Farid Azizi Jalilian
Gene Reports.2021; 23: 101107. CrossRef - Bacteriophage Therapy for Critical and High-Priority Antibiotic-Resistant Bacteria and Phage Cocktail-Antibiotic Formulation Perspective
Gursneh Kaur, Ritika Agarwal, Rakesh Kumar Sharma
Food and Environmental Virology.2021; 13(4): 433. CrossRef - A comprehensive review of bacterial osteomyelitis with emphasis on Staphylococcus aureus
Ahmad Nasser, Taher Azimi, Soheila Ostadmohammadi, Samaneh Ostadmohammadi
Microbial Pathogenesis.2020; 148: 104431. CrossRef - Characterization of the Three New Kayviruses and Their Lytic Activity Against Multidrug-Resistant Staphylococcus aureus
Natalia Łubowska, Bartłomiej Grygorcewicz, Katarzyna Kosznik-Kwaśnicka, Agata Zauszkiewicz-Pawlak, Alicja Węgrzyn, Barbara Dołęgowska, Lidia Piechowicz
Microorganisms.2019; 7(10): 471. CrossRef


 PubReader
PubReader Cite
Cite




