Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 4(5); 2013 > Article
-
Original Article
Evaluation and Selection of Multilocus Variable-Number Tandem-Repeat Analysis Primers for GenotypingBrucella abortus Biovar 1 Isolated from Human Patients - Subok Lee, Kyu-Jam Hwang, Mi-Yeoun Park, Seon-Do Hwang, Hee-Youl Chai, Hyuk Chu, Sang-Hee Park
-
Osong Public Health and Research Perspectives 2013;4(5):265-270.
DOI: https://doi.org/10.1016/j.phrp.2013.09.005
Published online: October 31, 2013
Division of Zoonoses, Korea National Institute of Health, Osong, Korea
- ∗Corresponding author. park3hee@korea.kr
© 2013 Published by Elsevier B.V. on behalf of Korea Centers for Disease Control and Prevention.
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Objectives
- Brucellosis is the most common bacterial zoonosis in the world. Multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) is a molecular method for genotyping bacterial species. Brucella abortus biovar I was isolated from most of the brucellosis-suspected patients in Korea. This study was conducted to investigate the ability of various MLVA primers that are used for molecular typing B. abortus isolates and for analyzing their epidemiological data.
-
Methods
- A total of 80 human isolates of B. abortus biovar I isolated from human patients and the reference strain were used for MLVA. Genetic diversity was determined by calculating the Simpson's diversity index (DI) of each VNTR locus. The Brucella strains were subcultured 30 times to determine the stability of each locus. The DNA of the strains cultivated in each passage was extracted and subjected to MLVA for further investigation.
-
Results
- The 15 VNTR loci were selected based on high DI values. The DIs of the 15 VNTR loci showed considerable discrimination power ranging from 59% for Bruce 43 to 87% for Bruce 22. Bruce 09, Bruce 11, Bruce 16, Bruce 42, and Bruce 43 were confirmed to remain stable in vitro among the 15 VNTR loci selected.
-
Conclusion
- The results of this study suggest that the five loci subsets may be a useful epidemiological tool for investigating B. abortus biovar 1 outbreak.
- Brucellosis, caused by species of the Gram-negative bacterium Brucella, continues to be a problem in humans and animals throughout the world [1]. In Korea, brucellosis is an endemic disease, and Brucella abortus is the prevailing strain in human infections [2]. Human brucellosis occurs in livestock workers and veterinarians who live and work with cattle farms in rural areas [3].
- Brucellosis spreads through foods contaminated with the bacterium Brucella, such as untreated milk products, cheese, or meat, as well as during contact with infected animals [4] and exposure to air containing the pathogen. Laboratory technicians and butchers usually acquire the infection when exposed to an infectious environment. It has been reported that more than 90% of brucellosis cases worldwide are attributed to Brucella melitensis and that B. abortus has been identified as the main pathogen causing the infection in Korea [5].
- Species identification and subtyping are important factors for epidemic surveillance and for investigating regions with a high incidence of brucellosis [6,7]. In general, microbiological methods and molecular typing are used to identify Brucella pathogens and to classify their species. B. abortus has been known to have at least seven biovars, and biovar 1 was isolated from most brucellosis-suspected patients in Korea. Phenotypic characterization does not provide sufficient data to trace the origin of the source of infection for epidemiological investigation. Recently, molecular biological methods have been developed and used for Brucella molecular typing purposes and epidemiological applications. This method is fast and easily standardized, and allows for strain clustering based on variable-number tandem-repeat (VNTR) similarities [8].
- In addition, because brucellosis has more than 90% DNA homology among species [8,9], epidemic studies have been mainly conducted using molecular genetic analysis [10]. Recent proactive studies on the genomic sequence of brucellosis and multilocus VNTR analysis (MLVA) have been mainly conducted for the identification of various Brucella species involved in causing the infection [9,11–14]. The MLVA has been used not only for Brucella identification but also for the identification of other pathogens [15]. Furthermore, it is useful for investigating molecular genetic pathogenesis.
- In this study, 78 MLVA primers were applied to 80 strains of B. abortus isolated from humans between 2003 and 2007. This study was conducted to identify those MLVA primers that are appropriate for the Korean isolates, to establish a method that rapidly identifies species using the selected primers, and to present epidemiological data for the molecular typing of B. abortus biovar 1 isolates.
Introduction
- 2.1 Bacterial isolates
- A total of 80 human brucellosis isolates were submitted to our laboratory in Korea during the period between 2003 and 2007 from the following ten provinces: Gyeonggi-do (GG), Gyeongbuk (GB), Gyeongnam (GN), Chungbuk (CB), Chungnam (CN), Daegu (DG), Jeonbuk (JB), Gwangju (GJ), Jeju (JJ), and Gangwon (GW). The reference strain, B. abortus 2308, was also included in the study. Genomic DNA was extracted using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany) by following the manufacturer's instructions.
- 2.2 In vitro stability test
- The stability of MLVA loci was evaluated in vitro using culture-passage methods. A single colony was taken from each of the B. abortus 2308 strain and human isolates 1/3, and inoculated on the blood agar plate. The strains were incubated for 3–4 days at 5% CO2, 37 °C for 30 passages. The DNA of the strains cultivated in each passage was extracted and subjected to MLVA.
- 2.3 Polymerase chain reaction amplification and electrophoresis
- The MLVA primers used in our assay were previously reported by Al Dahouk et al [8]. The DNA amplification was performed using 15 μL of solution containing 10 ng of genomic DNA, 1 U of i-Max II DNA polymerase (iNtRON Biotechnology, Gyeonggi-do, Korea), 2.5 mM deoxyribonucleotide triphosphate mixture, 10× polymerase chain reaction (PCR) buffer, 10 pmol of each primer (Bioneer, Daejeon, Korea), 5× betaine (Sigma Aldrich, St. Louis, MO, USA), and distilled sterilized water. Each reaction mixture was run in the Gene Amp PCR system 9700 (Applied Biosystems). After initial denaturation for 5 minutes at 96 °C, 30 cycles of multiple passages were ran: first, for a 30-second duration at 96 °C, followed by another 30-second run at 60 °C and a 1-minute run at 70 °C, with a final extension step at 70 °C for 5 minutes. The PCR product was loaded onto 2% or 3% agarose gel (GenDEPOT, Barker, NY, USA) with a size marker (100-bp ladder; Bioneer, Daejeon, Korea). Electrophoretic separation was run for 45 minutes.
- 2.4 Genotyping assay
- Brucella DNA was prepared using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany). The fluorescent primers were synthesized with one of the following three dyes (ABI, London, UK): NED (yellow), 6-FAM (blue), and HEX (green; Table 1). The PCR was performed using i-Max II polymerase (iNtRON Biotechnology) for DNA amplification. The PCR product was analyzed with an ABI PRISM 3730xl Analyzer using the ABI PRISM BigDye Terminator Cycle Sequencing kit containing AmpliTaq DNA polymerase.
- 2.5 Cloning and sequencing analysis
- The PCR product was cloned into a pCR 2.1-TOPO (Invitrogen, Carlsbad, CA, USA), and the plasmid was transformed into the competent Escherichia coli strain DH5a. Recombinant bacteria were selected and identified by blue/white screening.
- 2.6 Data analysis
- The genetic diversity of VNTRs was calculated using online tools [16]. The diversity index (DI) is a measure of variations in the number of repeats at each locus. It ranges from 0.0 (no diversity) to 1.0 (complete diversity).
Materials and Methods
- 3.1 Selection of MLVA primers appropriate for Korean Brucella subtyping
- The MLVA standardization was conducted on the reference strain, B. abortus 2308, and on the 80 human isolates of biovar 1, which were identified in the following Korean regions between 2003 and 2007: Gyeonggi (GG), Gyeongbuk (GB), Gyeongnam (GN), CB, CN, DG, JB, GJ, JJ, and GW. In this study, MLVA primers that are appropriate for Korean Brucella subtyping were selected using the 78 MLVA primers, including MLVA-16. The 78 primers were applied to the reference strain and the human isolates, and the results were shown as the size of the PCR product. When genetic diversity was measured using an online analysis tool, the DI was shown between 0% and 75%. Among the primers, Bruce 01, Bruce 14, Bruce 22, Bruce 43, Bruce 71, and Bruce 72 had a high diversity (Figure 1). In addition, Bruce 22, Bruce 01, Bruce 14, Bruce 43, and Bruce 71 had a Simpson DI of more than 0.5. The nonspecific band size was shown in Bruce 06. In the case of Bruce 15, however, it was difficult to discriminate among the various Korean isolates because there were no data on PCR amplification. The highest diversity was seen in 14 MLVA primers, including Bruce 01, Bruce 04, Bruce 07, Bruce 09, Bruce 11, Bruce 14, Bruce 16, Bruce 22, Bruce 25, Bruce 30, Bruce 42, Bruce 43, Bruce 71, and Bruce 72. These primers seemed to be effective in Korean Brucella subtyping.
- For further accurate analysis of 14 MLVA primers that were found to be appropriate for Korean Brucella subtyping, fluorescent-labeled genotyping was conducted using the 14 primers. Fluorescent labels, such as HEX, 6-FAM, and NED, were attached to the primers according to the size of PCR products (Table 1), and then the primer set was applied to the reference strain, B. abortus 2308, and the 80 human isolates. The genotyping data were analyzed using V-DICE software. The results of genotyping analysis were compared with those of electrophoresis analysis. More diverse discrimination was seen in genotyping using fluorescence labels. This may be attributed to the fact that a small difference that is difficult to be highly discriminated by electrophoresis analysis can be discriminated by fluorescent labeling. A diversity of ≥2 was observed in Bruce 2, Bruce 01, Bruce 14, Bruce 72, Bruce 71, Bruce 43, and Bruce 25 compared with that of electrophoresis analysis.
- To examine the accuracy of genotyping analysis, 12 PCR products that had a size different from that of the reference strain, B. abortus 2308, were selected among MLVA PCR products of the isolates, and their nucleotide sequence was analyzed. As a result of nucleotide sequence analysis, 11 of 12 cases were consistent with those of fluorescent-labeling analysis, and two cases were consistent with those of electrophoresis analysis. This result suggested that fluorescent labeling could be used for accurate analysis.
- 3.2 Genomic stability of multiple passages
- A previous study reported that the VNTR number of strains changes if multiple passages are performed [2]. Thus, the reference strain, B. abortus 2308, and human isolates 1/3 were examined after 30 cycles of multiple passages with intervals of 2–3 days. The DNA was then extracted from each strain, and mixed with the selected 14 primers to investigate consistency in PCR products. The change in PCR product size between the reference strain and human isolates 1/3 was observed in the ninth passage for Bruce 01, Bruce 04, Bruce 07, Bruce 14, Bruce 22, Bruce 25, Bruce 30, Bruce 71, and Bruce 72 among the 14 MLVA primers (Figure 2). Meanwhile, genomic stability without size change was observed for Bruce 09, Bruce 11, Bruce 16, Bruce 42, and Bruce 43 despite the 30 cycles of multiple passages.
- 3.3 Establishment of the condition of fluorescence-labeling multiplex MLVA
- For the development of rapid and easy MLVA, primer sets that are appropriate for multiplex PCR were designed to be used in electrophoresis. Two primer sets were prepared using the finally selected five primers to allow discrimination in genotyping without the overlapping of fluorescent labeling, and then multiplex PCR was conducted. When electrophoresis was performed, the difference in the PCR product size was found with naked eyes. Meanwhile, accurate discrimination was observed in genotyping analysis. Thus, genotyping analysis was confirmed as an analysis method that can identify primers for discrimination among Korean Brucellosis cases.
Results
- Methods for investigating pathogenesis have been significantly developed. Currently, MLVA has been mainly used for tracking the pathogenesis of bacterial genus with a high homology, such as the Brucella genus [11]. Because this method is applied to animals or humans to identify causative factors of infections where origin tracking of infection is difficult, that is, in cases involving infection of veterinarians or laboratory technicians and infection caused by food intake, appropriate molecular genetic analysis is required for tracking the route of transmission in Korea. Therefore, this study was conducted to investigate the origin tracking of endemic strains of biovar 1 human isolates collected from seven regions between 2003 and 2007. Based on a previous study indicating that discrimination among strains and origin tracking can be conducted using MLVA-16 that consist of three panels, the total MLVA, including the aforementioned MLVA-16, was applied to Korean human isolates. As a result, Bruce 01, Bruce 04, Bruce 07, Bruce 09, Bruce 11, Bruce 14, Bruce 16, Bruce 22, Bruce 25, Bruce 30, Bruce 42, Bruce 43, Bruce 71, and Bruce 72 were shown to be the MLVA primers that can discriminate between the reference strain, B. abortus 2308, and the isolates. These primers were also found to have a high diversity. The DI at 14 VNTR loci was shown to vary from 59% in Bruce 43 to 87% in Bruce 22 (Table 2). The 14 primers with a high diversity seem to be useful for discriminating between the reference strain and the Korean isolates and for the identification of the possible route of Brucella transmission.
- It has been known that changes in repeat number by multiple passages induce DNA insertion, deletion, and DNA point mutation. To assess the usefulness of the selected 14 primers, the reference strain, B. abortus 2308, and the isolates 1/3 underwent 30 cycles of multiple passages, and then the DNA of each strain was extracted. The obtained DNA was mixed with the primers selected from subtyping, followed by PCR. As a result, despite the 30 cycles of multiple passages, Bruce 09, Bruce 11, Bruce 16, Bruce 42, and Bruce 43 showed genomic stability without changes in the size of PCR product (Figure 2). Fluorescent-labeling multiplex PCR was performed to develop a rapid and accurate MLVA using the selected five primers. As a result, a small difference that is hard to be discriminated by electrophoresis was identified by this method. Because molecular genetic analysis requires the accuracy and stability of genetic markers, this rapid and accurate analysis method could be useful for molecular genetic analysis.
- In summary, of the 78 MLVA primers used in this study, Bruce 09, Bruce 11, Bruce 16, Bruce 42, and Bruce 43 allowed discrimination among the Korean isolates. Thus, a rapid and easy identification of strains was feasible by multiplex PCR using the five loci primers. In addition, these primers can be used for the identification of strains that cause an endemic outbreak and for tracking their origin, as well as for the identification of an outbreak of Brucella abortus isolates.
Discussion
-
Acknowledgements
- This research was supported by a fund (Grant Nos. 2010-N52002-00 and 2011-N52003-00) from Korea Centers for Disease Control and Prevention.
Acknowledgments
-
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Article information
- 1. Pappas G., Akritidis N., Bosilkovski M., Tsianos E.. Brucellosis. N Engl J Med 352(22). 2005 Jun;2325−2336. PMID: 15930423.ArticlePubMed
- 2. Park S.-H., Lee Y.-H., Chu H., Hwang S.-D., Choi H.-Y., Park M.-Y.. Application of the microagglutination test for serologic diagnosis of human brucellosis. Osong Public Health Res Perspect 3(1). 2012 Mar;19−23. PMID: 24159482.ArticlePubMed
- 3. Lee K., Lim H.S., Park W.W., Kim S.H., Lee D.Y., Park M.Y., Hur Y.. Seroprevalence of brucellosis among risk population in Gyeongsangbuk-do, 2006. J Prev Med Public Health 40(4). 2007 Jul;285−290. PMID: 17693731.ArticlePubMed
- 4. Tiller R.V., De B.K., Boshra M.. Comparison of two multiple-locus variable-number tandem-repeat analysis methods for molecular strain typing of human Brucella melitensis isolates from the Middle East. J Clin Microbiol 47(7). 2009 Jul;2226−2231. PMID: 19439543.ArticlePubMed
- 5. Park M.Y., Lee C.S., Choi Y.S., Park S.J., Lee J.S., Lee H.B.. A sporadic outbreak of human brucellosis in Korea. J Korean Med Sci 20(6). 2005 Dec;941−946. PMID: 16361801.ArticlePubMed
- 6. Al Dahouk S., Hagen R.M., Nockler K.. Failure of a short-term antibiotic therapy for human brucellosis using ciprofloxacin. A study on in vitro susceptibility of Brucella strains. Chemotherapy 51(6). 2005 Oct;352−356. PMID: 16227689.ArticlePubMed
- 7. Kattar M.M., Jaafar R.F., Araj G.F.. Evaluation of a multilocus variable-number tandem-repeat analysis scheme for typing human Brucella isolates in a region of brucellosis endemicity. J Clin Microbiol 46(12). 2008 Dec;3935−3940. PMID: 18923007.ArticlePubMed
- 8. Al Dahouk S., Flèche P.L., Nockler K.. Evaluation of Brucella MLVA typing for human brucellosis. J Microbiol Methods 69(1). 2007 Apr;137−145. PMID: 17261338.ArticlePubMed
- 9. Garofolo G., Ancora M., Di Giannatale E.. MLVA-16 loci panel on Brucella spp. using multiplex PCR and multicolor capillary electrophoresis. J Microbiol Methods 92(2). 2013 Feb 15;103−107. PMID: 23174277.Article
- 10. Whatmore A.M., Shankster S.J., Perrett L.L.. Identification and characterization of Variable-Number Tandem-Repeat Markers for typing of Brucella spp. J Clin Microbiol 44(6). 2006 Jun;1982−1993. PMID: 16757588.ArticlePubMed
- 11. Haguenoer E., Baty G., Pourcel C.. A multi locus variable number of tandem repeat analysis (MLVA) scheme for Streptococcus agalactiae genotyping. BMC Microbiol 11:2011 Jul;171PMID: 21794143.ArticlePubMed
- 12. Le Flèche P., Jacques I., Grayon M.. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol 6:2006 Feb;9PMID: 16469109.ArticlePubMed
- 13. Van Belkum A.. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA). FEMS Immunol Med Microbiol 49(1). 2007 Feb;22−27. PMID: 17266711.ArticlePubMed
- 14. Kulakov Y.K., Zheludkov M.M., Sclyarov O.D.. Variable-number tandem repeat markers for identification of Brucella abortus 82 and 75/79-AV vaccine strains. Vaccine 28(Suppl. 5). 2010 Oct;F41−F45. PMID: 20362200.ArticlePubMed
- 15. Rees R.K., Graves M., Caton N., Ely J.M., Probert W.S.. Single tube identification and strain typing of Brucella melitensis by multiplex PCR. J Microbiol Methods 78(1). 2009 Jul;66−70. PMID: 19410609.ArticlePubMed
- 16. http://www.hpa-bioinformatics.org.uk/cgi-bin/DICI/DICI.pl.
References
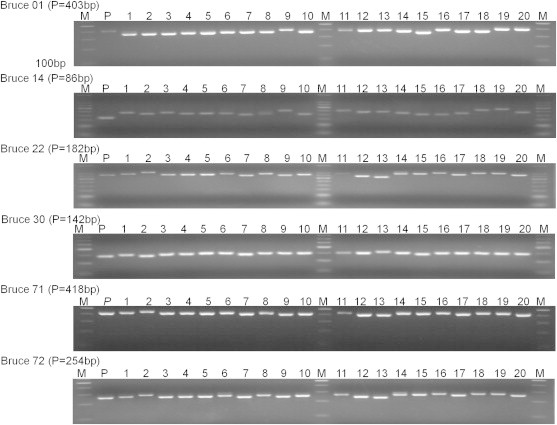
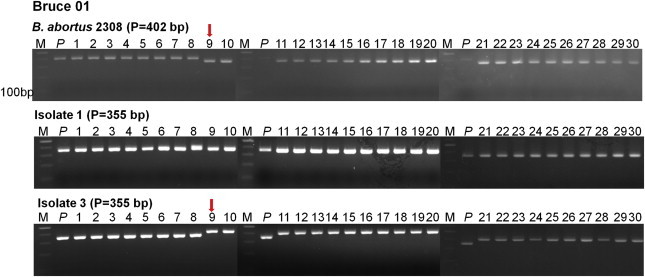
Figure & Data
References
Citations

- Brucella abortus: Current Research and Future Trends
Tariq Jamil, Falk Melzer, John Njeru, Hosny El-Adawy, Heinrich Neubauer, Gamal Wareth
Current Clinical Microbiology Reports.2017; 4(1): 1. CrossRef


 PubReader
PubReader Cite
Cite
