Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 14(6); 2023 > Article
-
Original Article
The value of CDC42 effector protein 2 as a novel prognostic biomarker in liver hepatocellular carcinoma: a comprehensive data analysis -
Hye-Ran Kim
 , Choong Won Seo
, Choong Won Seo , Jongwan Kim
, Jongwan Kim
-
Osong Public Health and Research Perspectives 2023;14(6):451-467.
DOI: https://doi.org/10.24171/j.phrp.2023.0229
Published online: December 15, 2023
Department of Biomedical Laboratory Science, Dong-Eui Institute of Technology, Busan, Republic of Korea
- Corresponding author: Jongwan Kim Department of Biomedical Laboratory Science, Dong-Eui Institute of Technology, 54 Yangji-ro, Busanjin-gu, Busan 47230, Republic of Korea E-mail: dahyun@dit.ac.kr
© 2023 Korea Disease Control and Prevention Agency.
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
- 893 Views
- 40 Download
Abstract
-
Objectives
- The prognostic significance of CDC42 effector protein 2 (CDC42EP2) and its association with tumor-infiltrating immune cells (TIICs) have not been explored in liver hepatocellular carcinoma (LIHC). This study aims to assess the potential prognostic value of CDC42EP2 by conducting a comprehensive analysis of online databases pertaining to LIHC.
-
Methods
- We evaluated the potential of CDC42EP2 as a prognostic biomarker by utilizing online databases such as TIMER, GEPIA2, KM, OSlihc, HPA, and LinkedOmics.
-
Results
- In LIHC, we observed that the mRNA and protein expression of CDC42EP2 were upregulated compared to normal tissues. Upregulated CDC42EP2 expression was associated with a worse prognosis based on the clinicopathological characteristics of patients with LIHC. Furthermore, CDC42EP2 was positively associated with TIICs. In the co-expression and functional enrichment analyses of CDC42EP2, 11,416 genes showed positive associations with CDC42EP2 while 8,008 genes showed negative associations. CDC42EP2-related co-expression genes were involved in protein localization to the endoplasmic reticulum, translational initiation, and RNA catabolic processes in gene set enrichment analysis-Gene Ontology (GSEA-GO), and regulated the ribosome, spliceosome, and primary immune deficiency in the GSEA-Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. In a survival map, 23 and 17 genes that exhibited positive associations with CDC42EP2 showed a significant hazard ratio (HR) for overall survival and disease-free survival, respectively.
-
Conclusion
- Our findings demonstrated that CDC42EP2 is a novel prognostic biomarker and a potential tumor immune therapeutic target in patients with LIHC.
- In 2020, liver cancer was identified as the third leading cause of cancer-related deaths worldwide, presenting a significant global health challenge. By 2025, it is estimated that there will be 1 million cases. Liver hepatocellular carcinoma (LIHC) is the most prevalent histological subtype of liver cancer [1]. LIHC ranks among the most common malignant tumors globally and is the primary cause of death in patients with cirrhosis [2,3]. It is associated with well-defined risk factors, such as hepatitis B and C viruses, excessive alcohol consumption, metabolic syndrome, and diabetes [4]. Over the past decade, significant progress in biochemical, pathological, and technical methods has enhanced the early diagnosis and treatment of LIHC, leading to better survival rates. These advancements have shed light on the molecular pathogenesis of LIHC; however, the therapeutic options currently available are still limited. The cure rate for LIHC is disappointingly low due to its aggressive nature, high recurrence rate, increased risk of metastasis, and poor response to chemotherapy. Additionally, achieving an early, definitive diagnosis of LIHC is often difficult. The majority of patients present with advanced-stage LIHC and have a dismal prognosis [5]. Despite recent progress in understanding the molecular etiology of LIHC, which has led to the development of new approved drugs, treatment options for advanced stages are scarce. Consequently, there is a pressing need for novel therapies for LIHC. The development of a new prognostic biomarker is crucial to enhance early diagnosis and improve the survival rates of patients with LIHC. In recent years, tumor immunotherapy has emerged as a promising new therapeutic strategy for LIHC.
- The immune system plays a crucial role in controlling cancer progression [6]. A previous study has demonstrated that tumor-infiltrating immune cells (TIICs) can help the host resist the development of cancer cells and solid tumors [7]. TIICs are a significant focus in cancer research [8]. The density and type of TIICs have a strong correlation with clinical outcomes of tumors and the efficacy of immunotherapy [9–12]. Some studies have highlighted the characteristics of the immune response and its link to prognosis [13,14]. The prognostic importance of TIICs and immune molecules, such as tumor-associated dendritic cells, macrophages, and natural killer cells, has been particularly noted in LIHC [15–17]. The TP53 mutation is common across various cancers and is associated with clinical prognostic outcomes [18,19]. However, the mechanisms by which TP53 mutations influence the relationship between CDC42 effector protein 2 (CDC42EP2) and TIICs remain unclear. CTNNB1 mutations occur in about 19% to 26% of LIHC patients, and these mutations are linked to immunological exclusion. Yet, systematic studies of TIICs in LIHC with CTNNB1 mutations and the mechanism by which these mutations cause immunological exclusion have not been conducted. In this context, we examined the correlation between CDC42EP2 and TIICs based on the mutation statuses of TP53 and CTNNB1 in LIHC. Copy number alterations (CNAs) represent one of the most frequent genetic variations in the human genome and are key molecular mechanisms in the pathogenesis of human diseases [20]. They can lead to the activation of oncogenes and the inactivation of tumor suppressor genes across various cancers. Consequently, CNAs are implicated in the pathogenesis of a wide range of cancers and play a crucial role in the molecular mechanisms of autoimmune and infectious diseases [21,22]. CNAs are also considered prognostic biomarkers for numerous diseases [23–26]. Understanding the mechanisms underlying TIIC-related prognoses and CNAs in LIHC is essential. However, the specific mechanisms connecting TIIC-related prognoses and CNAs in LIHC have yet to be elucidated. Therefore, the discovery of novel biomarkers associated with TIICs in LIHC could be instrumental in enabling early detection through the identification of specific immune mechanisms.
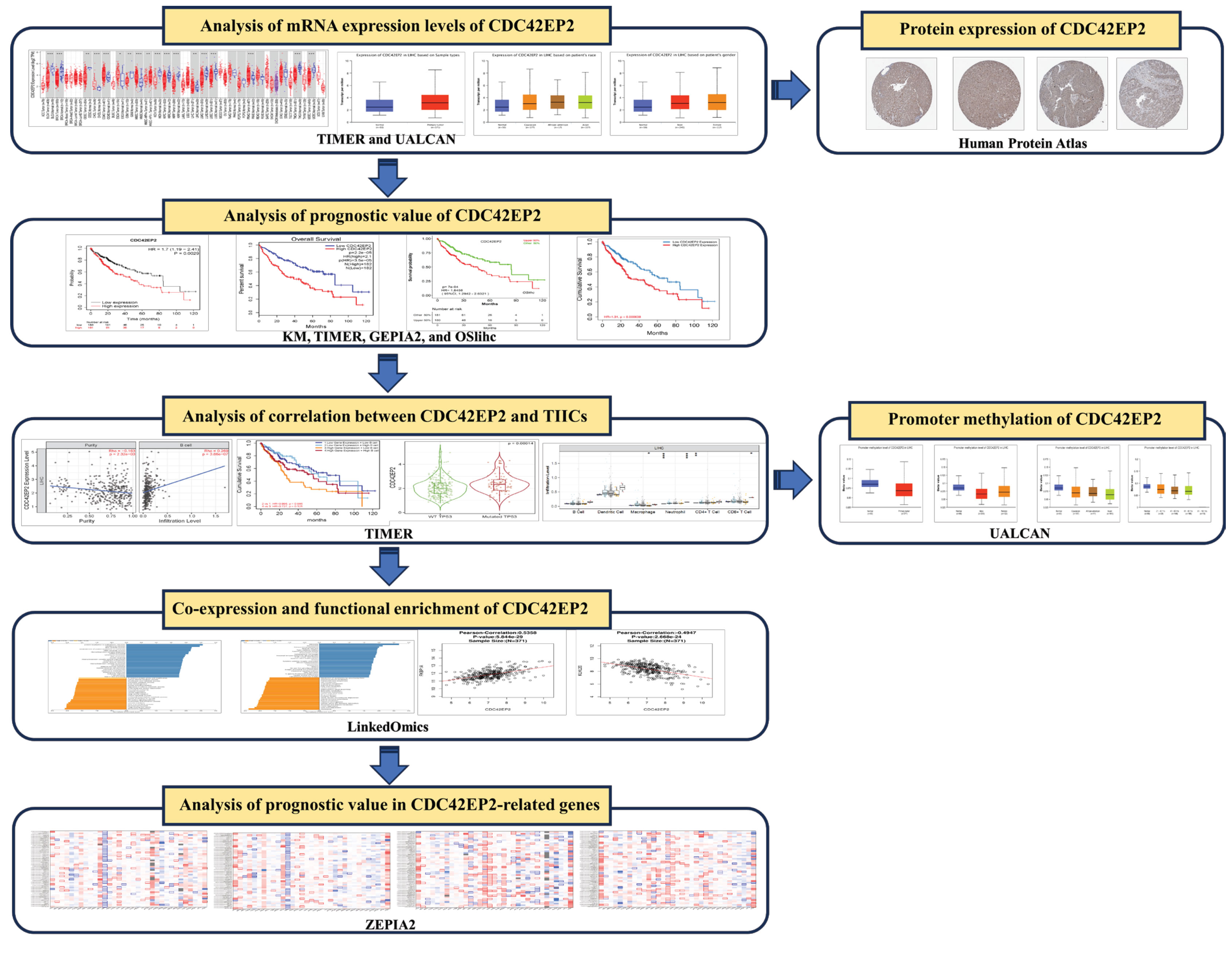
- In the present study, we aimed to evaluate the potential prognostic value of CDC42EP2 by analyzing its associations with clinicopathological features and TIICs in LIHC. We examined CDC42EP2 expression and its prognostic implications in LIHC using publicly available databases, including the Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/), UALCAN (http://ualcan.path.uab.edu), Gene Expression Profiling Interactive Analysis 2 (GEPIA2, http://gepia2.cancer-pku.cn/#index), Kaplan-Meier (KM, http://kmplot.com) plotter, and OSlihc (http://bioinfo.henu.edu.cn/DatabaseList.jsp). Additionally, we evaluated CDC42EP2 protein levels in liver cancer through the Human Protein Atlas (HPA, https://www.proteinatlas.org/) database. Employing the TIMER database, we explored the relationship between CDC42EP2 and TIICs, as well as the prognostic significance of their combined expression in LIHC. We also examined the correlations between TIICs and CDC42EP2-related gene mutations, as well as between TIICs and CNAs. Promoter methylation changes across various clinicopathological characteristics were investigated using the UALCAN database. The LinkedOmics (http://www.linkedomics.org/) database facilitated our analysis of CDC42EP2’s biological functions and co-expression patterns in LIHC. Furthermore, we utilized the GEPIA2 database to evaluate the prognostic relevance of genes associated with CDC42EP2 in a range of cancer types, including LIHC. Our research indicates that CDC42EP2 may serve as a viable target for therapeutic intervention and immunotherapy in LIHC patients. We aim to establish its utility as a prognostic biomarker for this disease.
Introduction
- TIMER Database Analysis
- TIMER is an online tool for analyzing immune cell infiltration across various cancer types. It leverages data from 10,897 samples within The Cancer Genome Atlas (TCGA) to estimate the abundance of immune infiltrates [27]. Our analysis focused on the expression and survival rates associated with CDC42EP2, as well as its clinical data. We examined the relationship between CDC42EP2 expression and TIICs, along with the prognostic significance of this association. Additionally, we explored the correlations between gene mutations (specifically TP53 and CTNNB1) and TIICs, as well as the relationships among CNAs, TIICs, and LIHC.
- UALCAN Database Analysis
- UALCAN is a database that utilizes TCGA level 3 RNA sequencing and clinical data from 31 cancer types. It enables users to analyze the relative expression of specific genes across both tumor and normal samples, as well as within various tumor subgroups. These subgroups can be categorized based on a range of clinicopathological features, including tumor stage, grade, race, sex, histological subtype, age, nodal metastasis, and TP53 mutation status [28]. Additionally, UALCAN offers tools to examine gene methylation levels. We analyzed the expression levels of CDC42EP2 in both tumor and normal tissues and explored the clinicopathological significance of CDC42EP2, along with its promoter methylation profiles. To assess the relationship between CDC42EP2 expression and promoter methylation, we utilized the plot module for cancer genomics, employing both Spearman and Pearson tests for our evaluation.
- Immunohistochemistry Staining Analysis
- HPA is a database that provides information on the distribution of proteins across various human tissues and cells [29]. For our analysis of CDC42EP2 proteomic expression levels, we obtained immunohistochemistry (IHC) images from the HPA specific to LIHC. We categorized the proteomic expression level of CDC42EP2 as “not detected,” “low,” “medium,” or “high.” This classification was determined by assessing the staining intensity and the proportion of cells that were stained.
- KM Plotter Database Analysis
- The KM plotter was used to estimate the effect of 54,000 genes on the prognosis of 21 common types of cancer [30,31]. This database comprises gene chip and RNA sequencing data sourced from repositories such as the Gene Expression Omnibus. Consequently, we assessed the prognostic significance of CDC42EP2 expression in patients with LIHC. Hazard ratios (HRs) with 95% confidence intervals (CIs) and log-rank p-values were calculated. A p-value of less than 0.05 was considered to indicate a statistically significant difference.
- OSlihc Database Analysis
- The OSlihc database serves as a platform for researchers to discover new prognostic biomarkers and may provide the opportunity to create novel targeted therapies for various cancers. Survival outcomes, such as overall survival (OS), disease-free interval (DFI), the progression-free interval (PFI), and disease-specific survival (DSS) were obtained to evaluate the prognostic value of CDC42EP2 in OSlihc [32].
- LinkedOmics Database Analysis
- LinkedOmics is a publicly accessible portal that contains data for 32 cancer types from TCGA [33]. It offers a distinctive resource for biologists and clinicians to explore multi-omics data related to cancer. The Kyoto Encyclopedia of Genes and Genomes (KEGG) is an online resource designed for the systematic analysis of gene functions and genomic information. Gene Ontology (GO) analysis categorizes gene functions into terms related to cellular components (CC), biological processes (BP), and molecular functions (MF). This categorization aids researchers in examining gene expression information from a network perspective. Pathway analyses facilitated by these tools provide detailed insights into signal transduction, transport, metabolism, and various other cellular activities. We conducted analyses of GO-BP and KEGG pathways using the “LinkInterpreter” module [33]. The ranking criterion was set at a false discovery rate of less than 0.05, and we carried out 500 simulations.
- GEPIA2 Analysis
- GEPIA2 was used to integrate clinical data from 9,736 tumors and 8,587 normal tissues derived from the TCGA and GTEx projects. The “Expression Analysis” module facilitated the conduction of clinical staging and survival analysis. We produced KM curves for OS and disease-free survival (DFS) associated with CDC42EP2 across 33 types of cancer, including LIHC, using the median value as the cut-off. Additionally, GEPIA2 offers a heat map that displays the survival analysis outcomes for various cancer types [34]. The survival results are presented as KM curves, accompanied by HRs and p-values obtained from the log-rank test. The threshold for significance in the Student t-test was set at a p-value of 0.05.
- Statistical Analysis
- For our analysis of gene expression data, we utilized the TIMER and UALCAN databases. We employed online tools such as the KM plotter, TIMER, OSlihc, and GEPIA2 to generate KM curves. The survival outcomes are presented as HRs with p-values from the log-rank test. A p-value of less than 0.05 from the log-rank test was considered to indicate a significant difference in survival times. The TIMER database was also used to assess the correlation between gene expression levels and immune signature scores, applying Spearman’s correlation coefficients. All data were sourced from publicly accessible databases, and the analyses were conducted using online tools. All reported results include p-values derived from the log-rank test, with values below 0.05 deemed to indicate statistical significance.
Materials and Methods
- mRNA Expression Analysis of CDC42EP2 in LIHC
- Figure 1 presents a flowchart of this study. To assess the differential expression of CDC42EP2 between tumor and normal tissues, we analyzed CDC42EP2 levels in various cancer types, including LIHC, using the TIMER database. We found that CDC42EP2 expression was elevated in LIHC, bladder cholangiocarcinoma, colon adenocarcinoma, esophageal carcinoma, head and neck squamous cell carcinoma (HNSC), prostate adenocarcinoma, thyroid carcinoma, and uterine corpus endometrial carcinoma when compared to normal tissues. Conversely, CDC42EP2 expression was reduced in bladder urothelial carcinoma (BLCA), breast invasive carcinoma, cervical squamous cell carcinoma and endocervical adenocarcinoma, glioblastoma multiforme, kidney chromophobe, kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, THCA, and UCEC relative to normal tissues, as shown in Figure 2A. We also examined the relationship between CDC42EP2 expression and various clinicopathological features in LIHC, including race, sex, histological subtype, age, stage, grade, lymph node metastasis status, and TP53 mutations. Our findings revealed a significant association between CDC42EP2 expression and primary tumor, race (Caucasian and Asian), sex, histological subtype (LIHC, hepatocholangiocarcinoma [HCC-CC]), age, grade (II, III, IV), stage (I, II, III), and TP53 mutations in LIHC. However, there was no correlation between CDC42EP2 expression and lymph node metastasis in LIHC, as depicted in Figure 2B. In summary, our data indicate that CDC42EP2 expression is upregulated in LIHC.
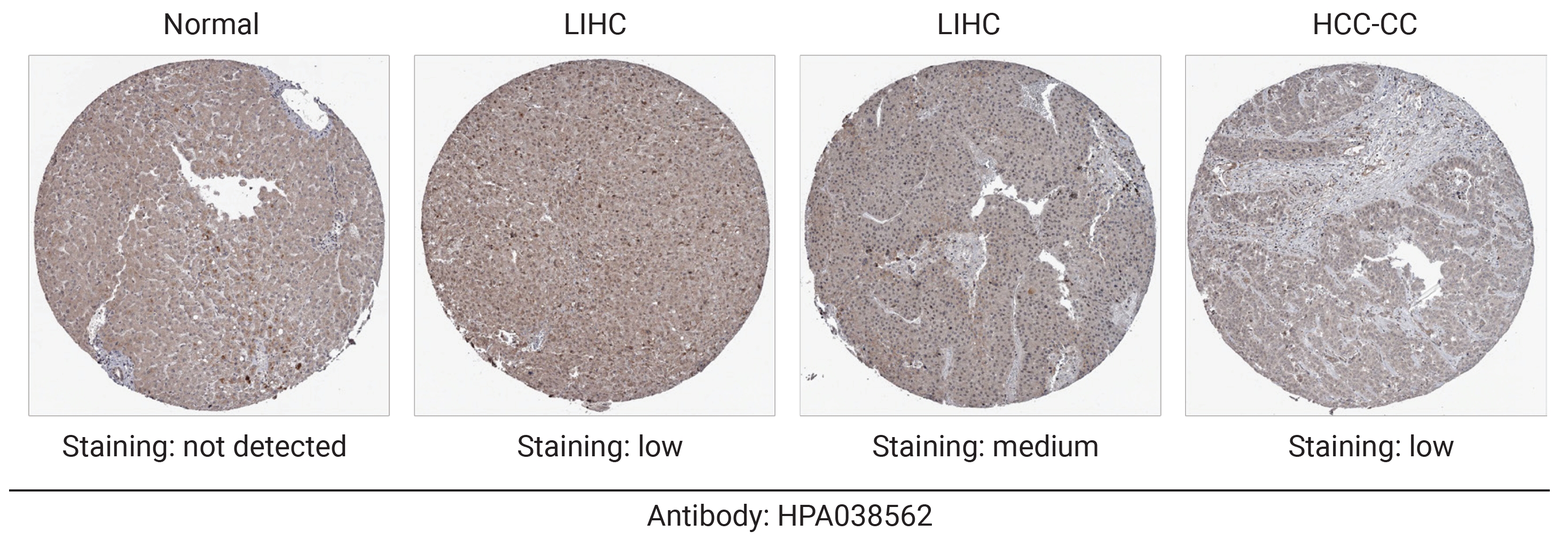
- Protein Expression of CDC42EP2 in LIHC
- To evaluate CDC42EP2 expression at the protein level, we analyzed CDC42EP2 protein expression levels using the HPA database. IHC images from the HPA indicated that CDC42EP2 protein expression was not detectable in normal liver tissue. In contrast, CDC42EP2 protein expression was significantly elevated in LIHC tissues compared to normal tissues. Additionally, low levels of CDC42EP2 were observed in HCC-CC (Figure 3). Our findings demonstrate that CDC42EP2 is overexpressed at both the transcriptional and translational levels in patients with LIHC.
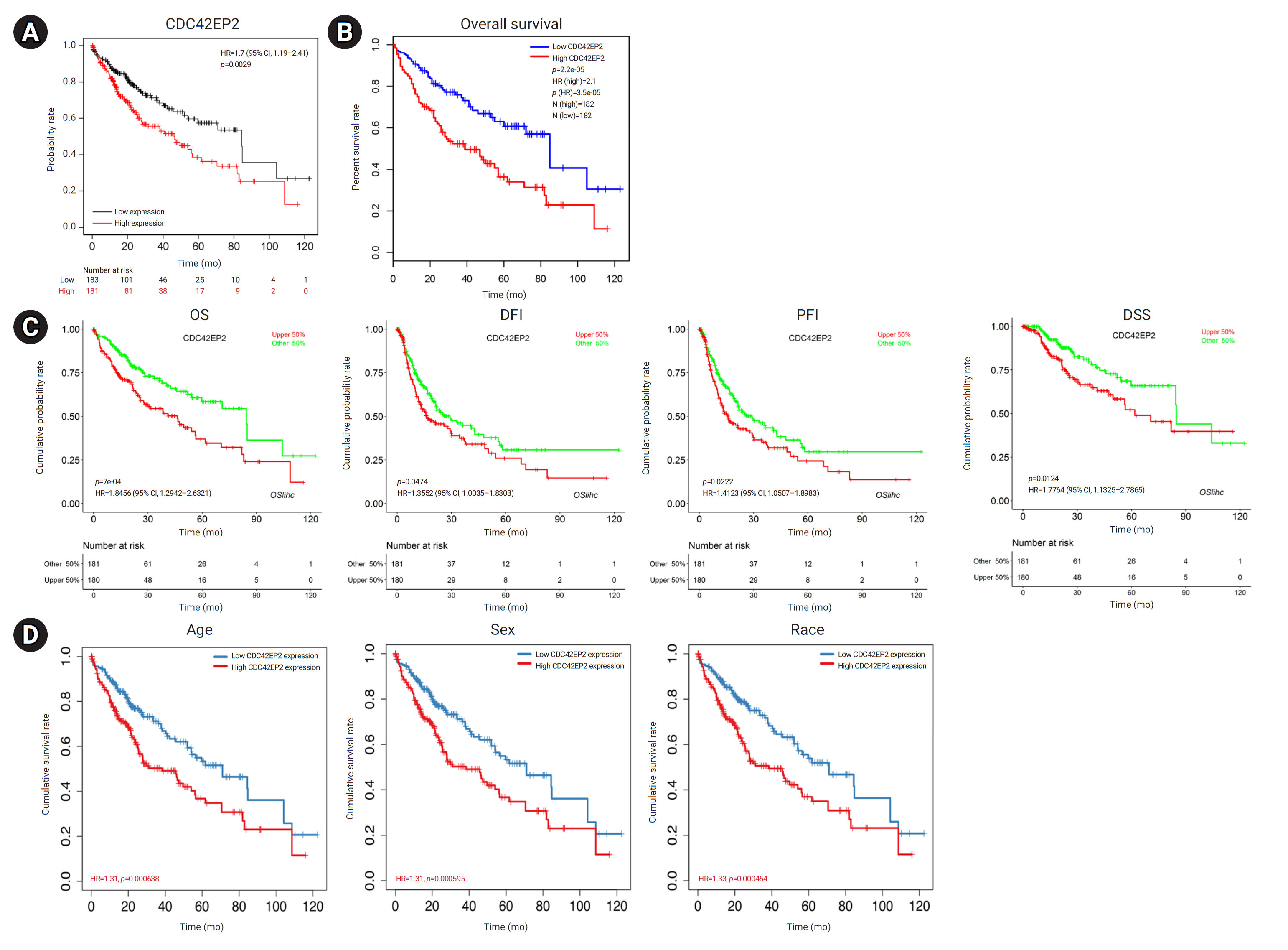
- Prognostic Value of CDC42EP2 Expression in LIHC
- To investigate the potential prognostic value of CDC42EP2 expression, we utilized the KM plotter, TIMER, GEPIA2, and OSlihc databases. Survival rates, including OS, DFI, PFI, and DSS, were analyzed according to the expression of CDC42EP2 in LIHC cells. Our findings indicated that increased CDC42EP2 expression was associated with significantly reduced OS (KM plotter: HR, 1.7; p=0.0029) (Figure 4A) (GEPIA2: HR, 2.1; p=0.000022) (Figure 4B). Furthermore, elevated CDC42EP2 expression was linked to poor prognosis in LIHC, as evidenced by OS (HR, 1.85; p=0.0007), DFI (HR, 1.36; p=0.0474), PFI (HR, 1.41; p=0.0222), and DSS (HR, 1.78; p=0.0124) (Figure 4C). Additionally, increased CDC42EP2 expression was associated with poor prognosis in LIHC across various demographics, including age (HR, 1.31; p=0.000638), sex (HR, 1.31; p=0.000595), and race (HR, 1.33; p=0.000454), as well as DSS (HR, 1.35; p=0.000488) (Figure 4D). To further confirm the prognostic significance of CDC42EP2 in LIHC, we examined the relationship between CDC42EP2 expression and the clinicopathological characteristics of LIHC patients using the OSlihc database. The clinicopathological features are depicted in Figure S1. The results demonstrated that increased CDC42EP2 expression was associated with poorer OS in both males (HR, 1.76; p=0.0141) and females (HR, 1.92; p=0.027), as well as in Asians (HR, 3.62; p=0.0002) and Caucasians (HR, 1.65; p=0.0351). Additionally, elevated CDC42EP2 expression was linked to worse prognosis in stage II (HR, 3.74; p=0.0047), stage III (HR, 2.52; p=0.0035), grade II (HR, 1.83; p=0.0228), and grade III (HR, 2.24; p=0.0132) LIHC. Increased CDC42EP2 expression also correlated with poorer DFI in Asians (HR, 1.57; p=0.0035) and PFI in females (HR, 1.73; p=0.039) and Asians (HR, 2.09; p=0.0026). Moreover, it was associated with worse PFI in Asians (HR, 4.65; p=0.0011), stage II (HR, 6.33; p=0.0168), stage III (HR, 2.10; p=0.0492), and grade III (HR, 3.17; p=0.0066). Furthermore, increased CDC42EP2 expression correlated with poor prognosis in BLCA, HNSC, and uveal melanoma (Figure S2). Taken together, our results show that upregulated CDC42EP2 expression predicts a poor prognosis of LIHC.
- Analysis of the Correlations between CDC42EP2 Expression and TIICs in LIHC
- Subsequently, we focused on correlations between CDC42EP2 expression and TIICs in LIHC using the TIMER database. CDC42EP2 was positively correlated with the infiltration levels of B cells (r=0.269, p=0.000000386), dendritic cells (r=0.443, p<0.0000000000000001), macrophages (r=0.261, p=0.000000845), neutrophils (r=0.075, p=0.148), CD4+ T cells (r=0.229, p=0.0000174), and CD8+ T cells (r=0.15, p=0.00529) in LIHC (Figure 5A). Next, we explored the association between CDC42EP2 expression, prognosis, and TIICs in LIHC. Our findings indicated that high CDC42EP2 expression combined with low B cell infiltration was linked to a poorer prognosis compared to low CDC42EP2 expression with low B cell infiltration. Similarly, high CDC42EP2 expression with low dendritic cell infiltration was associated with a poorer prognosis than low CDC42EP2 expression with low dendritic cell infiltration. A worse prognosis was also observed with high CDC42EP2 expression and high macrophage infiltration compared to low CDC42EP2 expression and low macrophage infiltration. Additionally, high CDC42EP2 expression with high neutrophil infiltration levels was linked to a poorer prognosis than low CDC42EP2 expression with low neutrophil infiltration. For CD4+ T cells, high CDC42EP2 expression with low infiltration was associated with a worse prognosis than low CDC42EP2 expression with high infiltration. Finally, high CDC42EP2 expression with low CD8+ T cell infiltration was linked to a poorer prognosis than low CDC42EP2 expression with low CD8+ T cell infiltration (Figure 5B). In summary, our results suggest that increased CDC42EP2 expression is associated with TIICs and may influence tumor prognosis in LIHC.
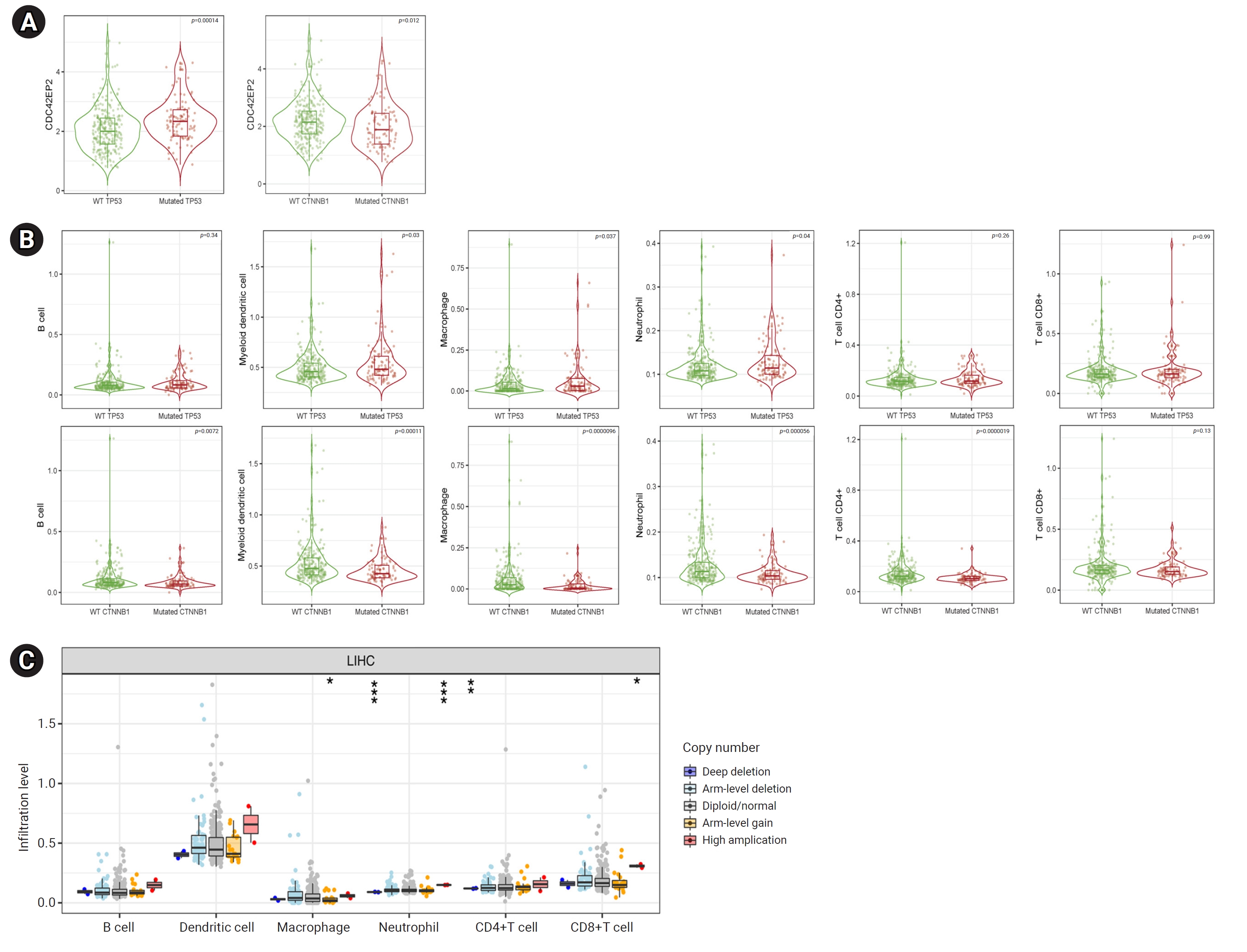
- Analysis of the Correlations between CDC42EP2 Expression and Gene Mutations in LIHC
- Correlations between gene mutations and CDC42EP2 expression in LIHC cells were investigated using the TIMER database. Increased expression of CDC42EP2 was observed in cells with TP53 (p=0.00014) and CTNNB1 (p=0.012) mutations (Figure 6A). We also examined the relationship between TP53 mutations and TIICs in LIHC patients. TP53 mutations were associated with higher levels of dendritic cells, macrophages, and neutrophils compared to the wild-type (WT) counterparts. Conversely, CTNNB1 mutations resulted in a reduced presence of B cells, dendritic cells, macrophages, neutrophils, and CD4+ T cells when compared to WT (Figure 6B). Additionally, we analyzed the TIICs in relation to CNAs of CDC42EP2 in LIHC. The findings indicated that CDC42EP2 CNAs were linked to changes in macrophages, neutrophils, and CD8+ T cells (Figure 6C). In summary, our data suggest a significant association between CDC42EP2 and mutations in TP53 and CTNNB1, as well as a notable correlation with the TIIC profiles of these mutations in LIHC.
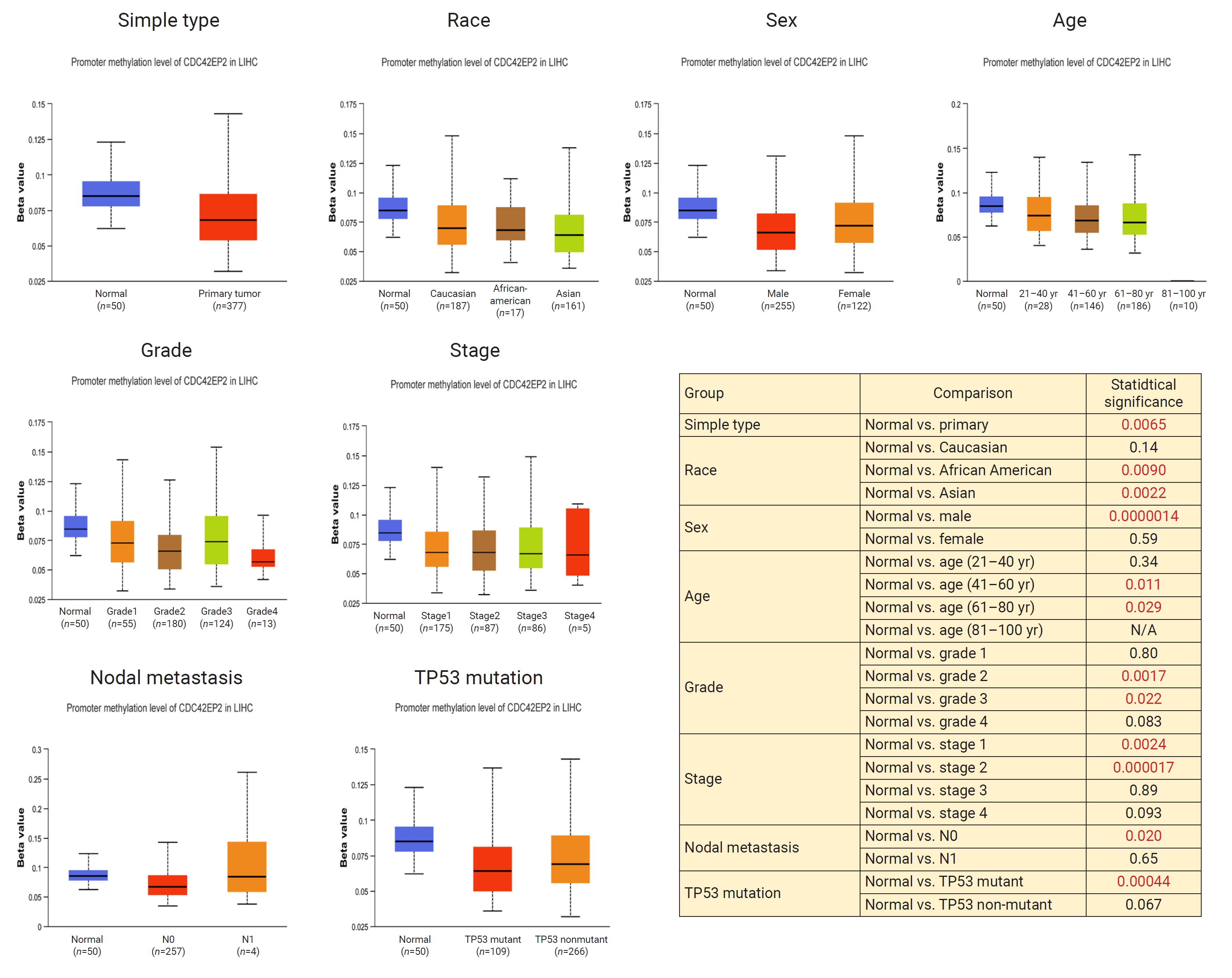
- Promoter Methylation Analysis for CDC42EP2 in LIHC
- We further investigated whether the expression of CDC42EP2 is affected by promoter methylation using the UALCAN platform. In LIHC, we found that CDC42EP2 expression is significantly associated with promoter hypomethylation in the primary tumor, race (African-American and Asian), sex (male), age groups (41–60 years, 61–80 years), tumor grade (II, III), tumor stage (I, II), and the presence of TP53 mutations. However, there was no significant relationship between promoter hypomethylation of CDC42EP2 and lymph node metastasis, as shown in Figure 7. These findings suggest that elevated expression of CDC42EP2 in LIHC may be a consequence of promoter hypomethylation.
- Co-Expression and Functional Enrichment Analysis of CDC42EP2 Expression in LIHC
- To explore the potential biological role and pathway of CDC42EP2 expression in LIHC, we assessed the co-expression of CDC42EP2 using the LinkedOmics database. A total of 11,416 genes demonstrated a positive association with CDC42EP2, indicated by dark red dots, while 8,008 genes exhibited a negative association, represented by dark green dots (Figure 8A). Heat maps were utilized to highlight the top 50 genes positively and negatively associated with CDC42EP2 (Figures 8B and C). The top five positively correlated genes included FKBP1A (r=0.5358, p=5.844e-29), EMP3 (r=0.5255, p=1.008e-27), LGALS2 (r=0.5212, p=3.201e-27), PLEKHO1 (r=0.5166, p=1.055e-26), and CMTM7 (r=0.5053, p=1.965e-25) (Figure 8D). Conversely, CDC42EP2 was negatively correlated with KLHL20 (r=–0.4947, p=2.668e-24), TOM1L1 (r=–0.488, p=1.365e-23), NNT (r=–0.4782, p=1.34e-22), C1orf25 (r=–0.4757, p=2.37e-22), and NFIC (r=–0.4648, p=2.773e-21) (Figure 8E). A comprehensive list of genes co-expressed with CDC42EP2 is provided in Table S1.
- We identified biological process categories using gene set enrichment analysis (GSEA) based on GO. The analysis revealed that genes co-expressed with CDC42EP2 are implicated in processes such as protein localization to the endoplasmic reticulum, translational initiation, and RNA catabolic processes (Figure 8F; Table S2). Additionally, a GSEA analysis of the KEGG pathways indicated that the co-expressed genes are predominantly enriched in ribosomes, spliceosomes, and primary immunodeficiency (Figure 8G; Table S3). In summary, our results suggest that CDC42EP2 may influence the prognosis of LIHC by modulating the global transcriptome.
- Prognostic Value of CDC42EP2-Related Genes in LIHC
- We investigated the prognostic value of CDC42EP2-associated genes in LIHC by analyzing data from the ZEPIA2 database. These CDC42EP2-associated genes appear to be high-risk factors for LIHC. Within the group of genes positively associated with CDC42EP2, 23 exhibited a high HR for OS (Figure 9A), while 17 demonstrated a high HR for DFS (Figure 9B). Conversely, among the genes negatively associated with CDC42EP2, MIA3 was associated with a low HR for OS (Figure 9C), and MYO18A was linked to a high HR for DFS (Figure 9D). CDC42EP2 was found to have a high HR in various cancer types, and genes negatively associated with CDC42EP2 generally showed a low HR across different cancers (Figure S3). Therefore, CDC42EP2 and its related genes are of prognostic importance in a range of cancers, including LIHC.
Results
- Liver cancer is a leading cause of mortality worldwide [35,36]. LIHC, the most common pathological form of liver cancer, often develops from chronic liver inflammation and fibrosis [37,38]. LIHC represents approximately 85% to 90% of liver cancer cases [35]. The prognosis for LIHC is extremely poor, largely because it is typically diagnosed at a late stage and there are limited therapeutic options available [37,39]. Therefore, there is an urgent need to identify useful biomarkers for the diagnosis and prognosis of LIHC, as well as treatment targets.
- The immune system plays a critical role in controlling cancer growth, and the activity of immune cells may actually promote cancer progression [6,40,41]. The tumor immune microenvironment (TIME), governed by TIICs, is a key player in cancer development and progression [42]. TIICs contribute to the evolution of cancer cells and tumors [7], and their quantity and type are directly linked to clinical outcomes [10,11]. Numerous studies have highlighted the characteristics of different immune cell types and their connections with prognosis [13,14], as well as the prognostic significance of TIICs and immune molecules in LIHC [15–17]. These findings underscore the importance of TIICs as critical prognostic markers and potential therapeutic targets. Therefore, TIICs may significantly influence the prognosis of LIHC patients. The TP53 gene is a renowned tumor suppressor involved in a range of anti-cancer processes, including apoptosis, senescence, cell cycle arrest, DNA repair, and the autophagy response [19]. Mutations in TP53 are the most common genetic alterations in human cancers, leading to uncontrolled cell proliferation and oncogenic activity [43,44], and are linked to poor outcomes in various cancer types [18,19,45]. The interplay between p53 mutations and immune system regulation has been extensively explored, particularly p53’s role in tumor immune regulation [46]. However, the mechanisms by which TP53 mutations affect the interaction between CDC42EP2 and TIICs remain unexplored. CTNNB1 is essential for the development of liver cancer and represents one of the most frequent mutations [47]. These mutations often occur at phosphorylation sites, leading to the accumulation of nuclear beta-catenin and the activation of the Wnt signaling pathway [48]. Mutations in CTNNB1 in LIHC are linked to immune exclusion and a poor prognosis. Yet, studies focusing on TIICs in the context of mutant CTNNB1 in LIHC are lacking. CNAs are among the most common genetic changes in the human genome and play a vital role in the molecular pathology of human diseases [20]. CNAs are associated with the activation of oncogenes and the suppression of tumor suppressor genes across various cancers, and they are instrumental in cancer development. Furthermore, CNAs have been identified as potential prognostic biomarkers in a range of diseases [23–26]. However, the specific mechanisms underlying TIIC-related prognoses and CNAs in LIHC remain unclear.
- In this study, we investigated the mRNA expression levels of CDC42EP2 in LIHC. We found that CDC42EP2 expression was upregulated in LIHC. Various clinicopathological factors were associated with this upregulation. While the CDC42EP2 protein was undetectable in normal tissues, its expression was significantly higher in LIHC tissues. Increased CDC42EP2 levels were linked to a poorer prognosis in LIHC, with this correlation being influenced by different clinicopathological factors. Our data strongly indicate that elevated CDC42EP2 could serve as a novel prognostic biomarker for LIHC. We also examined the relationship between CDC42EP2 and TIICs in LIHC. Our findings showed a positive correlation between CDC42EP2 expression and the presence of various TIICs, including B cells, dendritic cells, macrophages, neutrophils, CD4+ T cells, and CD8+ T cells. Moreover, increased CDC42EP2 levels were associated with a poorer prognosis. When analyzing TIICs in the context of CDC42EP2-related gene mutations and CNAs, we discovered that CDC42EP2 is linked to mutations in TP53 and CTNNB1, as well as to the TIICs associated with these mutations in LIHC. Additionally, CDC42EP2 significantly affects promoter hypomethylation in relation to clinicopathological factors. In investigating the co-expression and potential biological role of CDC42EP2 in LIHC, we identified 11,416 genes that showed a positive correlation with CDC42EP2, while 8,008 genes had a negative correlation. This suggests that CDC42EP2 has a widespread impact on the transcriptomic landscape of LIHC. This study also explored the prognostic significance of CDC42EP2-related genes in LIHC. Both CDC42EP2 and its related genes hold prognostic value in various cancer types, including LIHC. Numerous studies, including our own previous work, have confirmed the association between high gene expression and poor prognosis, echoing the findings of this study [49–53]. These collective results highlight the limitations of big data analyses and underscore the necessity for further research to understand the functional implications of these genes.
- In conclusion, our findings demonstrate that upregulated expression of CDC42EP2 correlates with poor prognosis and TIICs in LIHC. Consequently, CDC42EP2 represents a novel prognostic biomarker that offers insights into potential tumor immune therapeutic targets for LIHC patients. Further studies are required to investigate the detailed mechanisms of the CDC42EP2 gene as a potential prognostic biomarker through in vitro and in vivo research.
Discussion
- • Upregulated CDC42 effector protein 2 (CDC42EP2) is correlated with poor prognosis, depending on various clinicopathological factors in liver hepatocellular carcinoma (LIHC).
- • CDC42EP2 is positively associated with tumor-infiltrating immune cells (TIICs) such as B cells, dendritic cells, macrophages, neutrophils, CD4+T cells, and CD8+T cells in LIHC.
- • CDC42EP2 is associated with TP53 and CTNNB1 mutations and is related to TIICs of TP53 and CTNNB1 mutations in LIHC.
- • CDC42EP2 is a novel prognostic biomarker that provides insight into potential tumor immune therapeutic targets for patients with LIHC.
HIGHLIGHTS
Supplementary Material
Figure S1.
Figure S2.
Figure S3.
Table S3.
-
Ethics Approval
Not applicable. Written informed consent was obtained for publication of this study and accompanying images.
-
Conflicts of Interest
The authors have no conflicts of interest to declare.
-
Funding
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (RS-2022-00165637).
-
Availability of Data
All data generated or analyzed during this study are included in this published article. For other data, these may be requested through the corresponding author.
-
Authors’ Contributions
Conceptualization: JK; Data curation: JK; Formal analysis: JK; Investigation: HRK, CWS; Methodology: HRK; Project administration: HRK, CWS; Resources: HRK, CWS; Software: HRK; Supervision: HRK; Validation: HRK; Visualization: HRK; Writing–original draft: all authors; Writing–review & editing: all authors. All authors read and approved the final manuscript.
Article information




- 1. Villanueva A. Hepatocellular carcinoma. N Engl J Med 2019;380:1450−62.ArticlePubMed
- 2. Paul C, Khera L, Kaul R. Hepatitis C virus core protein interacts with cellular metastasis suppressor Nm23-H1 and promotes cell migration and invasion. Arch Virol 2019;164:1271−85.ArticlePubMedPDF
- 3. Sangiovanni A, Del Ninno E, Fasani P, et al. Increased survival of cirrhotic patients with a hepatocellular carcinoma detected during surveillance. Gastroenterology 2004;126:1005−14.ArticlePubMed
- 4. Singal AG, El-Serag HB. Hepatocellular carcinoma from epidemiology to prevention: translating knowledge into practice. Clin Gastroenterol Hepatol 2015;13:2140−51.ArticlePubMedPMC
- 5. Zheng X, Jin W, Wang S, et al. Progression on the roles and mechanisms of tumor-infiltrating T lymphocytes in patients with hepatocellular carcinoma. Front Immunol 2021;12:729705. ArticlePubMedPMC
- 6. Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015;21:938−45.ArticlePubMedPMCPDF
- 7. Bremnes RM, Al-Shibli K, Donnem T, et al. The role of tumor-infiltrating immune cells and chronic inflammation at the tumor site on cancer development, progression, and prognosis: emphasis on non-small cell lung cancer. J Thorac Oncol 2011;6:824−33.ArticlePubMed
- 8. Cabrita R, Lauss M, Sanna A, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020;577:561−5.ArticlePubMedPDF
- 9. Angell HK, Lee J, Kim KM, et al. PD-L1 and immune infiltrates are differentially expressed in distinct subgroups of gastric cancer. Oncoimmunology 2018;8:e1544442.ArticlePubMedPMC
- 10. Choi Y, Kim JW, Nam KH, et al. Systemic inflammation is associated with the density of immune cells in the tumor microenvironment of gastric cancer. Gastric Cancer 2017;20:602−11.ArticlePubMedPDF
- 11. Hao X, Luo H, Krawczyk M, et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci U S A 2017;114:7414−9.ArticlePubMedPMC
- 12. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541−50.ArticlePubMedPMCPDF
- 13. Sia D, Jiao Y, Martinez-Quetglas I, et al. Identification of an Immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology 2017;153:812−26.ArticlePubMed
- 14. Kurebayashi Y, Ojima H, Tsujikawa H, et al. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 2018;68:1025−41.ArticlePubMedPDF
- 15. Harding JJ, Khalil DN, Abou-Alfa GK. Biomarkers: what role do they play (if any) for diagnosis, prognosis and tumor response prediction for hepatocellular carcinoma? Dig Dis Sci 2019;64:918−27.ArticlePubMedPDF
- 16. Sun H, Huang Q, Huang M, et al. Human CD96 correlates to natural killer cell exhaustion and predicts the prognosis of human hepatocellular carcinoma. Hepatology 2019;70:168−83.ArticlePubMedPDF
- 17. Tian MX, Liu WR, Wang H, et al. Tissue-infiltrating lymphocytes signature predicts survival in patients with early/intermediate stage hepatocellular carcinoma. BMC Med 2019;17:106. ArticlePubMedPMCPDF
- 18. Zhang H, Huang Z, Song Y, et al. The TP53-related signature predicts immune cell infiltration, therapeutic response, and prognosis in patients with esophageal carcinoma. Front Genet 2021;12:607238. ArticlePubMedPMC
- 19. Zhang X, Cheng Q, Yin H, et al. Regulation of autophagy and EMT by the interplay between p53 and RAS during cancer progression (Review). Int J Oncol 2017;51:18−24.ArticlePubMed
- 20. Inaki K, Liu ET. Structural mutations in cancer: mechanistic and functional insights. Trends Genet 2012;28:550−9.ArticlePubMed
- 21. Beeghly-Fadiel A, Lu W, Shu XO, et al. MMP9 polymorphisms and breast cancer risk: a report from the Shanghai Breast Cancer Genetics Study. Breast Cancer Res Treat 2011;126:507−13.ArticlePubMedPMCPDF
- 22. Schofield JB, Krausz T, Stamp GW, et al. Ossifying fibromyxoid tumour of soft parts: immunohistochemical and ultrastructural analysis. Histopathology 1993;22:101−12.ArticlePubMed
- 23. Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics 2002;3:229−43.ArticlePubMed
- 24. Yu X, Huang J, Wu S, et al. Copy number variations of MMP-9 are prognostic biomarkers for hepatocellular carcinoma. Transl Cancer Res 2020;9:698−706.ArticlePubMedPMC
- 25. Xu W, Zhou W, Cheng M, et al. Hypoxia activates Wnt/β-catenin signaling by regulating the expression of BCL9 in human hepatocellular carcinoma. Sci Rep 2017;7:40446. ArticlePubMedPMCPDF
- 26. Kim TM, Yim SH, Shin SH, et al. Clinical implication of recurrent copy number alterations in hepatocellular carcinoma and putative oncogenes in recurrent gains on 1q. Int J Cancer 2008;123:2808−15.ArticlePubMedPMC
- 27. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017;77:e108. −10.ArticlePubMedPMCPDF
- 28. Wu Z, Xia C, Zhang C, et al. Prognostic significance of SNCA and its methylation in bladder cancer. BMC Cancer 2022;22:330. ArticlePubMedPMCPDF
- 29. Chen G, Luo D, Zhong N, et al. GPC2 is a potential diagnostic, immunological, and prognostic biomarker in pan-cancer. Front Immunol 2022;13:857308. ArticlePubMedPMC
- 30. Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725−31.ArticlePubMedPDF
- 31. Lanczky A, Nagy A, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat 2016;160:439−46.ArticlePubMedPDF
- 32. An Y, Wang Q, Zhang G, et al. OSlihc: an online prognostic biomarker analysis tool for hepatocellular carcinoma. Front Pharmacol 2020;11:875. ArticlePubMedPMC
- 33. Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46(D1):D956−63.ArticlePubMedPMC
- 34. Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47(W1):W556−60.ArticlePubMedPMCPDF
- 35. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394−424.ArticlePubMedPDF
- 36. Nguyen K, Jack K, Sun W. Hepatocellular carcinoma: past and future of molecular target therapy. Diseases 2015;4:1. ArticlePubMedPMC
- 37. Uehara T, Ainslie GR, Kutanzi K, et al. Molecular mechanisms of fibrosis-associated promotion of liver carcinogenesis. Toxicol Sci 2013;132:53−63.ArticlePubMedPMC
- 38. Dapito DH, Mencin A, Gwak GY, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012;21:504−16.ArticlePubMedPMC
- 39. Wallace MC, Preen D, Jeffrey GP, et al. The evolving epidemiology of hepatocellular carcinoma: a global perspective. Expert Rev Gastroenterol Hepatol 2015;9:765−79.ArticlePubMed
- 40. Ostrand-Rosenberg S. Immune surveillance: a balance between protumor and antitumor immunity. Curr Opin Genet Dev 2008;18:11−8.ArticlePubMedPMC
- 41. Munhoz RR, Postow MA. Recent advances in understanding antitumor immunity. F1000Res 2016;5:2545. ArticlePubMedPMCPDF
- 42. Lazar DC, Avram MF, Romosan I, et al. Prognostic significance of tumor immune microenvironment and immunotherapy: novel insights and future perspectives in gastric cancer. World J Gastroenterol 2018;24:3583−616.ArticlePubMedPMC
- 43. Xu F, Lin H, He P, et al. A TP53-associated gene signature for prediction of prognosis and therapeutic responses in lung squamous cell carcinoma. Oncoimmunology 2020;9:1731943. ArticlePubMedPMC
- 44. Long J, Wang A, Bai Y, et al. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine 2019;42:363−74.ArticlePubMedPMC
- 45. Jiang Z, Liu Z, Li M, et al. Immunogenomics analysis reveals that TP53 mutations inhibit tumor immunity in gastric cancer. Transl Oncol 2018;11:1171−87.ArticlePubMedPMC
- 46. Guo G, Yu M, Xiao W, et al. Local activation of p53 in the tumor microenvironment overcomes immune suppression and enhances antitumor immunity. Cancer Res 2017;77:2292−305.ArticlePubMedPMCPDF
- 47. Tornesello ML, Buonaguro L, Tatangelo F, et al. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics 2013;102:74−83.ArticlePubMed
- 48. Gao C, Wang Y, Broaddus R, et al. Exon 3 mutations of CTNNB1 drive tumorigenesis: a review. Oncotarget 2017;9:5492−508.ArticlePubMedPMC
- 49. Ma J, Jin J, Lu H, et al. Exonuclease 1 is a potential diagnostic and prognostic biomarker in hepatocellular carcinoma. Front Mol Biosci 2022;9:889414. ArticlePubMedPMC
- 50. Lu T, Li C, Xiang C, et al. Overexpression of CISD1 predicts worse survival in hepatocarcinoma patients. Biomed Res Int 2022;2022:7823191. ArticlePubMedPMCPDF
- 51. Xie S, Wang Y, Huang J, et al. A novel m6A-related prognostic signature for predicting the overall survival of hepatocellular carcinoma patients. IET Syst Biol 2022;16:1−17.ArticlePubMedPMCPDF
- 52. Kim HR, Seo CW, Han SJ, et al. C4orf47 is a novel prognostic biomarker and correlates with infiltrating immune cells in hepatocellular carcinoma. Biomed Sci Lett 2023;29:11−25.Article
- 53. Kim HR, Seo CW, Lee JH, et al. SAMD13 as a novel prognostic biomarker and its correlation with infiltrating immune cells in hepatocellular carcinoma. Biomed Sci Lett 2022;28:260−75.Article
References
Figure & Data
References
Citations




 Cite
Cite










