Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 14(5); 2023 > Article
-
Review Article
Strategies to combat Gram-negative bacterial resistance to conventional antibacterial drugs: a review -
Priyanka Bhowmik1
 , Barkha Modi2
, Barkha Modi2 , Parijat Roy1
, Parijat Roy1 , Antarika Chowdhury1
, Antarika Chowdhury1
-
Osong Public Health and Research Perspectives 2023;14(5):333-346.
DOI: https://doi.org/10.24171/j.phrp.2022.0323
Published online: October 18, 2023
1Department of Biological Sciences, School of Life Science & Biotechnology, Adamas University, Kolkata, India
2Department of Microbiology, Techno India University, Kolkata, India
- Corresponding author: Priyanka Bhowmik Department of Biological Sciences, School of Life Science & Biotechnology, Adamas University, Barrackpore-Barasat Road, 24 Paragnas North, Jagannathpur, Kolkata, West Bengal, India E-mail: pri.4383@gmail.com
© 2023 Korea Disease Control and Prevention Agency.
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Abstract
- The emergence of antimicrobial resistance raises the fear of untreatable diseases. Antimicrobial resistance is a multifaceted and dynamic phenomenon that is the cumulative result of different factors. While Gram-positive pathogens, such as methicillin-resistant Staphylococcus aureus and Clostridium difficile, were previously the most concerning issues in the field of public health, Gram-negative pathogens are now of prime importance. The World Health Organization’s priority list of pathogens mostly includes multidrug-resistant Gram-negative organisms particularly carbapenem-resistant Enterobacterales, carbapenem-resistant Pseudomonas aeruginosa, and extensively drug-resistant Acinetobacter baumannii. The spread of Gram-negative bacterial resistance is a global issue, involving a variety of mechanisms. Several strategies have been proposed to control resistant Gram-negative bacteria, such as the development of antimicrobial auxiliary agents and research into chemical compounds with new modes of action. Another emerging trend is the development of naturally derived antibacterial compounds that aim for targets novel areas, including engineered bacteriophages, probiotics, metal-based antibacterial agents, odilorhabdins, quorum sensing inhibitors, and microbiome-modifying agents. This review focuses on the current status of alternative treatment regimens against multidrug-resistant Gram-negative bacteria, aiming to provide a snapshot of the situation and some information on the broader context.
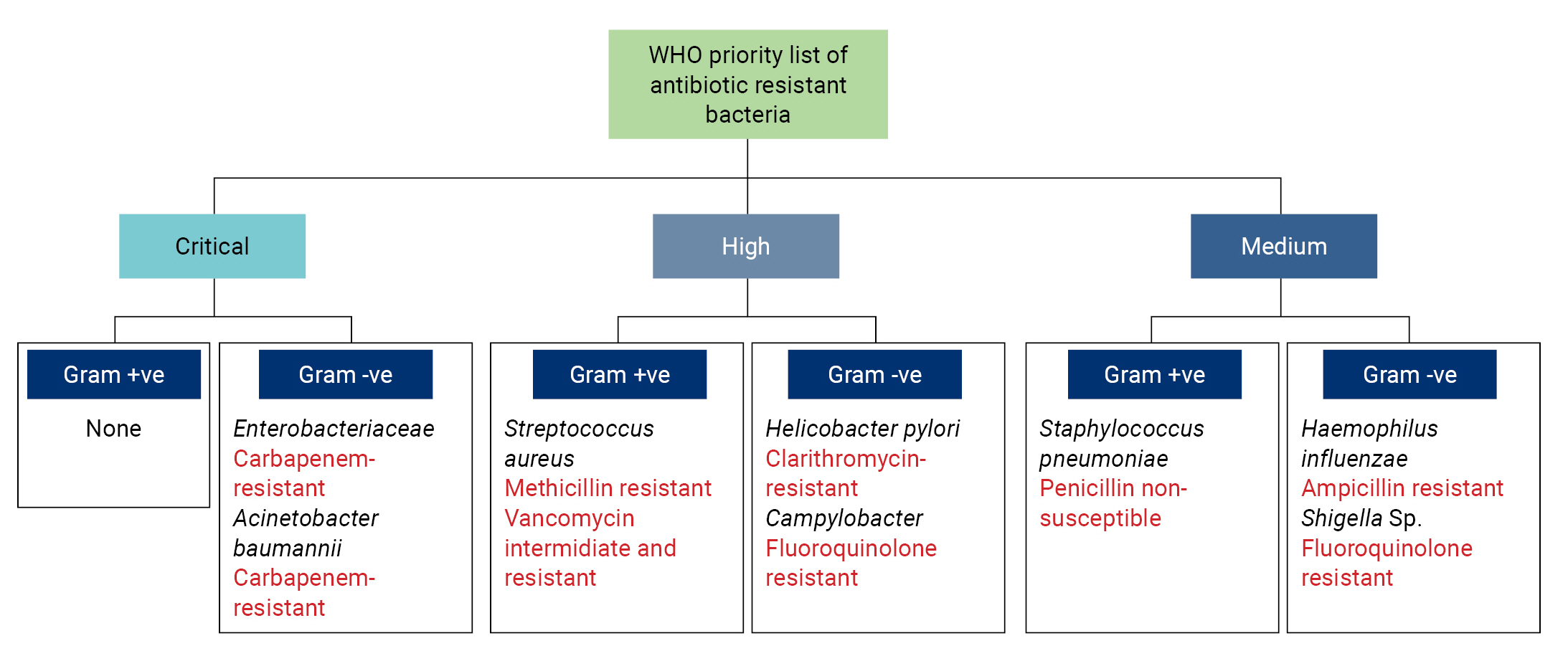
- Antibiotic resistance is a global public health threat caused by the imprudent use and widespread overdosing of antibiotics [1,2]. Antimicrobial resistance poses a significant challenge in clinical settings, leading to devastating infections. In 2017, the World Health Organization (WHO) released a list of priority pathogens (Figure 1) [3], for which new medicines are urgently needed. These pathogens were categorized as critical, high, or medium priority [4,5]. The majority of the pathogens listed by the WHO are Gram-negative bacteria. It has been clearly demonstrated that Gram-negative bacteria have a higher potential to cause serious diseases in humans, particularly among immunocompromised individuals, due to their specialized cellular structure [1,3]. The outer membrane (OM) of Gram-negative bacteria has been identified as the primary factor promoting resistance against a broad range of antibiotics. The OM functions as a permeability barrier, effectively blocking many current antibiotics, and it exerts its effect in 2 ways: by reducing influx and increasing efflux.
- Reduced Influx
- Membrane proteins permit the selective entry of various molecules. Numerous antibiotics, including penicillin, carbapenems, cephalosporins, and fluoroquinolones, gain access to the bacterial cell via these proteins. Changes in membrane permeability can influence the entry of these antibiotics, leading to resistance [6]. OM porin proteins are particularly significant in the context of antibiotic resistance, as they can limit the entry of several antibiotics, such as β-lactams and fluoroquinolones, into the cell [7,8].
- Increased Efflux
- Furthermore, even when an antibiotic successfully penetrates the OM, it can be swiftly expelled from the cell by a variety of broadly acting efflux pumps, thereby inducing resistance. These pumps utilize either the energy derived from ATP hydrolysis or the proton motive force to expel chemicals from the cell. For instance, an ABC (ATP-binding cassette) superfamily pump has been demonstrated to be linked with heightened resistance to aminoglycosides and polymyxins in Serratia marcescens [9].
- Efforts to combat clinically relevant drug resistance have involved modifying existing antibiotic classes or introducing new antibiotics. Following the initial “golden era” of antibiotics, large pharmaceutical companies began to encounter significant scientific challenges in their search for new antibiotics, particularly those effective against Gram-negative bacterial drug resistance. This led to a waning interest in a field that no longer promised ever-increasing profits. However, the emergence of extreme drug resistance has prompted global public health authorities to seek alternatives to antibiotics. In this review, we have categorized antibiotic substitutes into 2 broad groups: (a) antibiotic adjuvants and (b) antibiotic alternatives. Antibiotic alternatives are further classified into (1) phages or phage-derived proteins, (2) direct-acting compounds, (3) repurposed approved drugs, (4) anti-virulence therapies, (5) RNA-based therapeutics, (6) nanomaterial-based therapeutics, and (7) miscellaneous.
Introduction
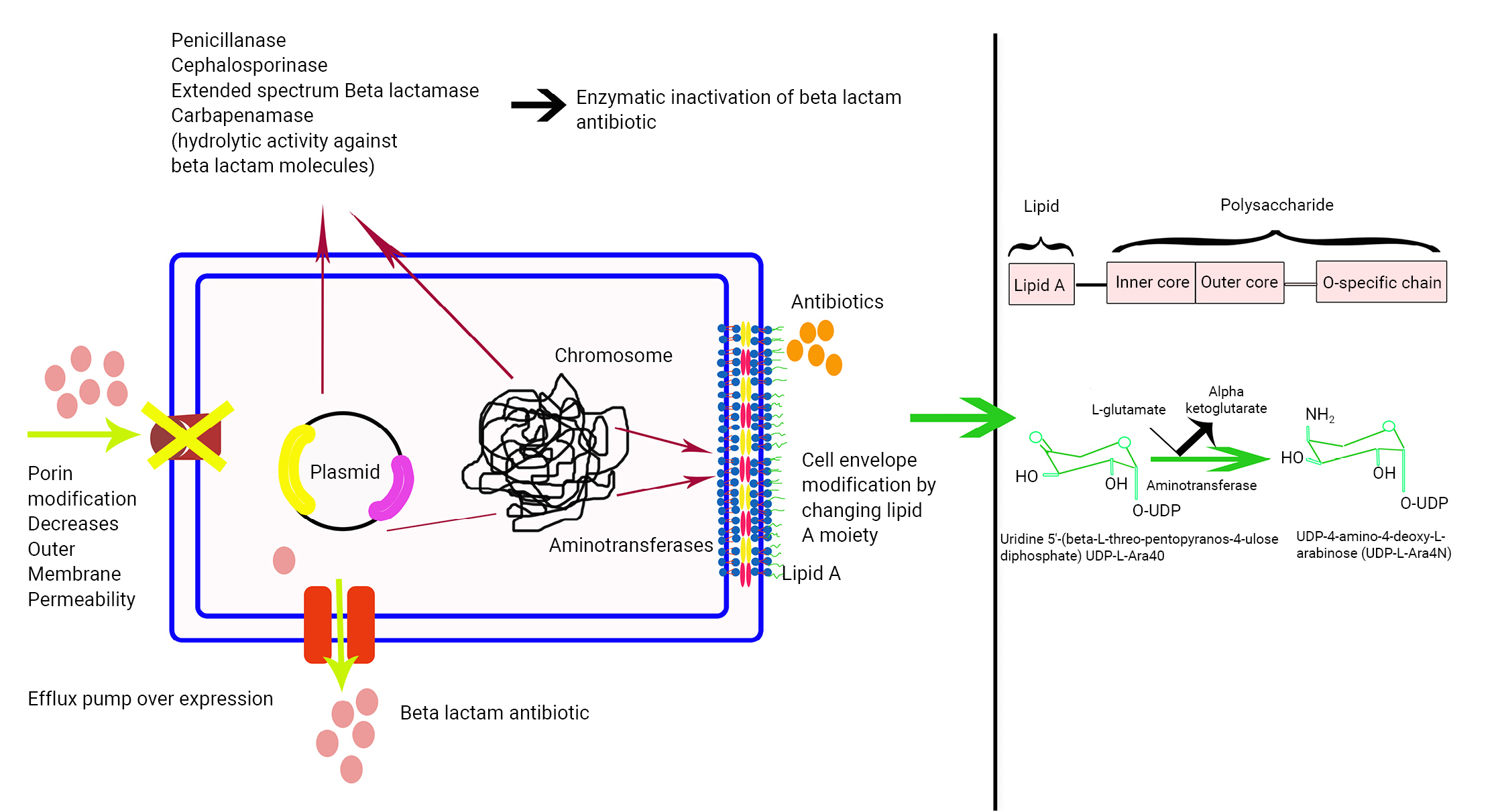
- In 2017, the WHO released a list of 12 antibiotic-resistant priority pathogens (Figure 1), which are considered to be the most significant threats to public health. All the critical priority list pathogens are Gram-negative bacteria, namely carbapenem-resistant Acinetobacter baumannii, carbapenem-resistant Pseudomonas aeruginosa, and carbapenem and third-generation cephalosporin-resistant Enterobacteriaceae (Escherichia coli, Enterobacter spp., and Klebsiella pneumoniae). Apart from these pathogens, clarithromycin-resistant Helicobacter pylori, fluoroquinolone-resistant Campylobacter sp. and Salmonella sp., and third-generation cephalosporin and fluoroquinolone-resistant Neisseria gonorrhoeae are also included in the WHO’s list of high-priority pathogens, along with Gram-positive Staphylococcus aureus and Enterococcus faecium [4,10]. Recently Gram-negative bacteria have also recently acquired resistance toward the last-resort drugs, polymyxins. The following section briefly discusses the different mechanisms of resistance. Figure 2 provides a visual representation of these resistance mechanisms.
- Mechanism of Resistance
- Bacteria employ different enzymes to exert resistance towards different antibiotics, either by degrading the antibiotics or by interfering with their activities. The mechanisms of action of these enzymes are shown in Figure 3.
- The β-lactam antibiotics hold prime importance. β-lactamases are enzymes that hydrolyze the amide bond of the 4-membered β-lactam ring, which leads to the inactivation of the antibiotic. The subsequent result is resistance.
- Carbapenem resistance: Resistance to carbapenems typically arises from 2 factors: (1) non-carbapenemase-mediated carbapenem resistance, and (2) the production of carbapenemases, which are enzymes that hydrolyze carbapenem antibiotics. The production of an extended-spectrum β-lactamase or AmpC enzymes, coupled with a decrease in cell membrane permeability due to modifications in porin proteins, has been reported to contribute to carbapenem resistance in Enterobacteriaceae [11]. The most common carbapenamase enzymes are K. pneumoniae carbapenemases (KPCs), which circulate in Enterobacteriaceae worldwide. Strains harboring KPCs are resistant to all β-lactams and often develop resistance to other classes of antibiotics, such as fluoroquinolones, aminoglycosides, and trimethoprim-sulfamethoxazole. The global prevalence of KPC-producing Enterobacteriaceae is increasing, leading to a worldwide multidrug resistance pandemic. OXA-48 and NDM are among the most prevalent types of carbapenemases found in K. pneumoniae [12]. The acquisition of carbapenemases has also been well-documented in A. baumannii [13].
- TEM, CTX-M, and extended-spectrum β-lactamase (ESBL)—cephalosporin resistance: These enzymes are broadly distributed via plasmids and other mobile genetic elements. The acquisition of point mutations in these enzymes allows them to hydrolyze oxyiminocephalosporins, such as cefotaxime and ceftazidime. This confers the so-called “extended-spectrum” phenotype, also known as ESBLs [14].
- The primary cause of resistance to polymyxins, specifically colistin, is generally attributed to chemical alterations of the lipid A moiety. These alterations result in a net decrease in negative charge, which in turn leads to a diminished affinity for the positively charged colistin molecule. The pmrHFIJKLM operon (also known as arnBCADTEF or pbgPE) has been identified as being associated with colistin-resistant bacteria. In a similar vein, plasmid-borne mcr genes, which possess phosphatidyl aminotransferase properties, facilitate the transfer of the phosphoethanolamine moiety to lipid A in the OM, thereby altering the structure of lipid A. Consequently, colistin is unable to penetrate into the periplasmic space, resulting in resistance to colistin [15].
- Porins are the most abundant proteins in the OM of Gram-negative bacteria. These water-filled open channels allow the selective entry of certain chemicals and are closely associated with resistance to various antibiotics. For instance, β-lactams and fluoroquinolones penetrate the cell through the non-specific porin OmpF [16,17]. Some Gram-negative bacteria with mutations in OmpF have been reported to be resistant to several β-lactam antibiotics, such as E. coli, K. pneumoniae, S. marcescens, P. aeruginosa, and Enterobacter aerogenes. Carbapenem sensitivity in A. baumannii is associated with the reduced expression of many porins, such as Caro and Omp [18]. Colistin tolerance in A. baumannii is promoted by the loss of lipopolysaccharide, as well as OM proteins, resulting in a reduction in membrane integrity [19].
- Antibiotic target sites, including penicillin-binding proteins and DNA gyrase, can undergo modifications. Alterations have been found in fluoroquinolone antibiotic targets such as parC and gyrA in P. aeruginosa isolates, which confer resistance to fluoroquinolone. A single point mutation, C257T, in the gyrA gene results in an amino acid substitution in the gyrase A subunit. This mutation has been identified in fluoroquinolone-resistant Campylobacter strains and appears to be horizontally transferred among these strains. The emergence of the fluoroquinolone-resistant Shigella sonnei population, which first appeared in South Asia, was facilitated by the sequential accumulation of mutations (gyrA-S83L, parC-S80I, and gyrA-D87G) [20]. In contrast, resistance to aminoglycosides, which exert their antibacterial effect by binding and inhibiting bacterial 16S ribosomal RNA (rRNA), emerges through modification of the antibiotic target 16S rRNA [21−24]. Mutations in genes such as pmrAB and phoPQ, which are associated with OM lipid A modification, can lead to colistin resistance in P. aeruginosa [25]. Clarithromycin resistance can develop in bacteria owing to mutations in the 23S rRNA gene, such as A2142G, A2142C, or A2143G. These mutations decrease the drug’s affinity, which is very important in the treatment of H. pylori infection [3,26].
- A high level of intrinsic resistance in Gram-negative bacteria is largely mediated by resistance-nodulation-division efflux pumps, which are capable of transporting antibiotics out of the bacterial cell. Various experiments have demonstrated that the inactivation of 1 or more components of efflux pumps is associated with increased susceptibility to antibiotics. High levels of efflux pump expression are associated with strains isolated in clinical settings. Evidence suggests that other resistance mechanisms also depend on efflux pump-mediated resistance. For example, the loss of efflux activity can alter the expression of other genes. The deletion of acrB or tolC has been correlated with decreased expression of the OM porin OmpF in Salmonella [27]. Additionally, the presence or absence of efflux can influence the rate at which antibiotic resistance mutations occur in a population [28,29].
Resistant Gram-Negative Bacteria and the Mechanisms of Their Resistance
Enzymes
β-Lactamases
Aminotransferase
Porin proteins
Target site modifications
Efflux pumps
- Antibiotic resistance is a grave and escalating issue. The Centers for Disease Control and Prevention report that antimicrobial resistance claims over 700,000 lives annually, a figure projected to surge to 10 million by 2055. Consequently, the urgent discovery of new antimicrobial drugs is deemed a top priority. Recent advancements have begun to concentrate more on natural products, reintroducing natural product screening in the quest for innovative therapeutics to tackle resistant bacterial diseases. In this section, we will explore several ground-breaking alternative strategies that have surfaced from research and development programs, specifically for combating drug-resistant Gram-negative bacteria.
- Antibiotic Adjuvants
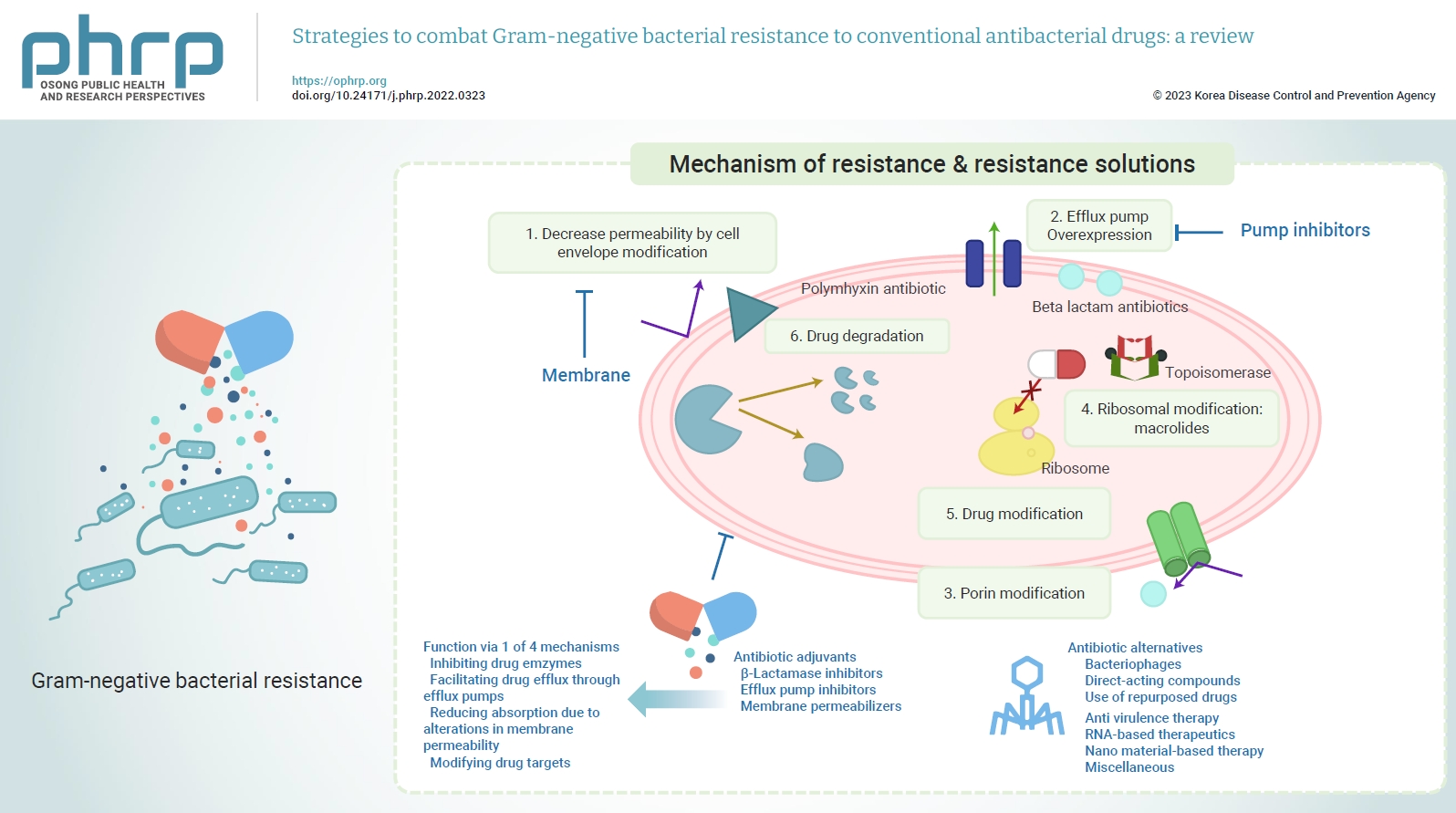
- Antibiotic adjuvants, also referred to as “resistance breakers” or “antibacterial potentiators,” are chemicals that possess little to no antibiotic activity on their own. However, when these adjuvants are combined with other antibiotics in a treatment regimen, they enhance the efficacy of the antibiotic. The use of antibiotic adjuvants has revitalized the application of several antibiotics against resistant organisms, thereby reducing the need for the discovery of new, complex, and costly antibiotics, as their efficacy can be restored simply by adding adjuvants [1].
- Thus far, 3 types of antibiotic adjuvants have been investigated or developed: lactamase antagonists, efflux pump antagonists, and OM permeabilizers (Table 1). These antibiotic adjuvants function via 1 of 4 mechanisms to aid antibiotics in overcoming bacterial resistance: inhibiting drug enzymes, facilitating drug efflux through efflux pumps, reducing absorption due to alterations in membrane permeability, and modifying drug targets [30].
- In clinical practice, the most commonly used antibiotic adjuvants are lactamase inhibitors [14,30]. They are employed to counteract resistance to β-lactam antibiotics [1]. Despite their long-term use, for over 70 years, β-lactamase inhibitors have been recognized as effective antibiotic adjuvants. These inhibitors work by hydrolyzing the amide bond of the 4-membered β-lactam ring in β-lactam antibiotics [1,14]. They can be divided into 2 distinct groups: (1) serine β-lactamases (SBLs) and (2) metallo-β-lactamases (MBLs). In SBLs, the lactamase’s nucleophilic serine forms a covalent bond with a hydrolyzed β-lactam. These are classified as Ambler categories A, C, or D. Enzymes in this group are inhibited by sulbactam, tazobactam, and clavulanic acid. In contrast, MBLs, which contain 1 or 2 active site zinc ions, attack β-lactams via polarized water molecules. MBLs are classified as B1, B2, or B3, based on the number of bonded zinc ions and sequence identity. MBLs are effective against all β-lactam antibiotics, except for monobactams. Currently, there are no approved inhibitors for class B MBLs [31−33]. β-lactamase inhibitors are typically administered in conjunction with β-lactam antibiotics to inhibit bacterial β-lactamases. Generally, the antimicrobial spectrum of these combinations is determined by the activity of the β-lactam and the characteristics of the β-lactamase inhibitor.
- Clavulanic acid is a β-lactam chemical produced by Streptomyces clavuligerus. It was discovered in 1976 that S. clavuligerus could produce clavulanic acid, making it the first agent used in conjunction with amoxicillin to inhibit β-lactamase. Structurally similar to penicillin, clavulanic acid forms a covalent bond with β-lactamase through a catalytic serine, creating a stable adduct. This binding occurs near the β-lactamase active site, effectively blocking enzymatic activity and enhancing the antibiotic’s effects. Currently, clavulanic acid is combined with amoxicillin and ticarcillin. However, it should be noted that clavulanic acid has no clinical effect against class B, C, and D β-lactamases.
- Sulfones based on penicillin, such as sulbactam and tazobactam, among others, are highly effective in combating antimicrobial resistance. Sulbactam is a semi-synthetic β-lactamase inhibitor. It binds irreversibly to a β-lactamase in proximity to its active site, thereby inhibiting these enzymes and preventing the degradation of β-lactam antibiotics.
- Tazobactam belongs to the class of penicillanic acids. It irreversibly attaches itself to the β-lactamase enzyme near its active site, thereby shielding β-lactam antibiotics from the enzyme’s activity [34,35].
- Zidebactam is a β-lactamase inhibitor classified as a bicyclo-acyl hydrazide [34,36]. Currently, WCK5222, a combination of zidebactam and cefepime, is undergoing phase 1 clinical trials investigating whether it could be used to treat severe infections caused by Gram-negative multidrug-resistant microorganisms [37].
- The target enzymes of β-lactam antibiotics facilitate transpeptidase and carboxypeptidase reactions during the biosynthesis of bacterial cell walls. Both the formation and deacylation of acyl-enzyme complexes in transpeptidase and SBL catalysis occur via a high-energy tetrahedral (sp3-hybridised) intermediate. Boronic acids and boronate esters, particularly cyclic ones, can effectively inhibit both SBLs and MBLs. They do this by rapidly reacting with SBLs and MBLs to form stable enzyme-inhibitor complexes that mimic the common high-energy tetrahedral intermediates in SBL/MBL catalysis [38].
- The cyclic boronate ester RPX7009 (vaborbactam), which has been shown to restore carbapenem activity against KPC25, is the most promising homologue to date [39]. Carbavance, a combination of RPX7009 and biapenem, is currently in phase 3 clinical trials for the treatment of KPC-producing Enterobacteriaceae infections [38]. Despite their effectiveness, no standalone (cyclic) boronates have been developed for clinical use that can compare to the currently used β-lactam antibiotics. However, there are reports suggesting that monocyclic vaborbactam and, in particular, bicyclic boronates (VNRX-5133 and related compounds) hold potential for use as broad-spectrum β-lactamase inhibitors when used in conjunction with suitable β-lactam antibiotics.
- Given the role of efflux pumps in antibiotic resistance, the importance of their inhibitors cannot be overstated. Most efflux pump inhibitors (EPIs) function by physically blocking substrate molecules from passing through the transporter. There are several EPIs commonly used in therapy, including reserpine, which is used to combat resistance to tetracycline and norfloxacin, and berberine, which is used to counteract imipenem resistance caused by P. aeruginosa [40]. Berberine, although not as effective alone, has been shown to potentiate ciprofloxacin against multidrug-resistant K. pneumoniae isolates [41]. PaβN also has shown broad effectiveness against antibiotic-resistant bacteria [42]. Alkylaminoquinazoline derivates have been found to be able to restore antibiotic activity in Gram-negative resistant isolates [43,44]. PAβN, 1-(1-naphthylmethyl)-piperazine (NMP) and carbonyl cyanide 3-chlorophenylhydrazone (CCCP) are the most studied synthetic EPIs studied with A. baumannii [43].
- Membrane permeabilizers increase the permeability of the Gram-negative OM, facilitating the increased entry of antibiotics. These substances can chelate and eliminate positively charged divalent ions from the OM, leading to a disruption in the structural organization of the OM [45].
- Antibiotic Alternatives
- Bacteriophages are bactericidal viruses that infect bacteria. Phage therapy, an alternative to antibiotics, is regaining interest worldwide due to the rise of broad-spectrum antibiotic resistance among various bacterial species. Lytic phages, which lyse their host bacterial cells, are particularly intriguing as potential antimicrobial agents. Numerous proteins assist in this process, including the widely studied holin protein. These small hydrophobic proteins create holes in the bacterial membrane, leading to a subsequent loss of proton motive force across the membrane. For instance, the S. aureus phage GH15 holin protein HolGH15 has demonstrated activity against Listeria monocytogenes in addition to S. aureus [46]. Another phage-encoded protein, endolysin, which is involved in peptidoglycan degradation, is also being explored as a potential antimicrobial agent. It has been reported that extracellularly supplied endolysin can cause lysis of the cell to which it has been applied, through a mechanism known as “lysis from without.” Many endolysins have shown broad-spectrum activity. For example, Salmonella phage BSPM4 M4Lys endolysin has demonstrated in vitro activity against numerous Gram-negative bacteria, such as Salmonella enterica, E. coli, P. aeruginosa, and Cronobacter sakazakii [47,48]. LysSAP26 endolysin can inhibit many drug-resistant bacteria, such as A. baumannii, E. coli, K. pneumoniae, P. aeruginosa, S. aureus, and E. faecium [49]. However, endolysin activity against Gram-negative bacteria is somewhat limited by the presence of an OM, which inhibits the synergistic effect of endolysin with antibiotics. A study demonstrated that the combination of Cp-711 with the β-lactam antibiotic amoxicillin or cefotaxime can be effective against drug-resistant Streptococcus pneumoniae. Endolysin LysECD7 was found to be effective against biofilms formed by K. pneumoniae [50]. Endolysins are also being used to create “enzybiotics,” which can be modified by adding extra domains to enhance their peptidoglycan-degrading ability. These fusions have shown activity against many Gram-negative pathogens [51]. The OM, which presents a significant resistance to phage endolysins, can be destroyed by another phage protein, called spanin. Holin, endolysin, and spanin together form the “lytic cassette.” As such, these proteins could also be exploited as potential tools against drug-resistant Gram-negative bacteria. Another protein, amurin, mimics the mode of action of β-lactam antibiotics and could be used as an alternative to antibiotics. While phage resistance in bacteria is not uncommon, phages can mutate and counter the phage resistance of bacteria.
- One benefit of utilizing phages as alternatives to antibiotics is their host specificity. Unlike antibiotics, phages do not harm the normal microflora. Additionally, phages do not carry the typical side effects associated with antibiotics, such as allergic reactions and anaphylactic shock.
- However, bacterial resistance to phages is inevitable. This resistance can hinder phage therapy due to the potential emergence of phage-resistant bacterial variants. A continuous “arms race” exists between the phage and its host, resulting in the co-evolution of both the virus and the bacteria. This co-evolution and resulting adaptability pose concerns for the development of phage therapy. However, the issue of phage resistance can be mitigated by using “phage cocktails,” which involve the use of more than 1 type of phage [52].
- The most recent clinical trial involving bacteriophages was a phase 1/2 randomized, controlled, double-blind study. The aim was to determine the effectiveness and tolerability of Phago-Burn, a combination of 12 naturally lytic P. aeruginosa bacteriophages in an alginate template, for treating P. aeruginosa-infected burn wounds [53,54]. In this trial, Phago-Burn was immediately applied to the wound. It was observed that the concentration of the phage cocktail had decreased after production, resulting in patients receiving a lower dosage than initially anticipated. Several commercial applications of phage therapy have been demonstrated, including Agriphage (Omnilytics Ltd.), Listex (Micreos Ltd.), SalmFresh (Intralytix Ltd.), ListShield (Intralytix Ltd.), and EcoShield (Intralytix Ltd.). Further research is required to establish a standard protocol that ensures public safety.
- Plants are rich sources of various bioactive molecules, such as polyphenols, phenols, terpenoids, numerous phytochemicals, and essential oils. Many of these have significant antimicrobial effects. For instance, allicin, derived from garlic, can inhibit a broad range of bacteria, including Escherichia (including enterohaemorrhagic E. coli 0157 and enterotoxigenic E. coli), Salmonella, Streptococcus, Staphylococcus, Klebsiella, Proteus, and H. pylori. Many plant-derived compounds have been reported to inhibit quorum sensing in bacteria, resulting in reduced motility and biofilm formation, factors directly related to antibiotic resistance. Many phytochemicals are currently being studied as potential alternatives to antibiotics, particularly in antibiotic-resistant bacteria, including multidrug-resistant strains [55−57]. Extracts from thyme, lemongrass, tulsi, neem, aloe vera, oregano, and rosemary have been shown to be effective against multidrug-resistant clinical isolates [58]. Tannins and flavonoids from plant extracts were even found to inhibit methicillin-resistant S. aureus (MRSA). Extracts of various plants like Lawsonia, Curcuma longa, Zingiber officinale, and Tinospora cordifolia were shown to be capable of inhibiting clinical isolates. The synergistic action of antibiotics and plant chemicals has also been proven to be effective against a range of resistant pathogens, supporting the use of plant-derived chemicals against drug-resistant bacteria [59]. Boswellic acid, derived from Boswellia sp., has been reported to be effective against Staphylococcus epidermidis, Enterococcus faecalis, and E. coli biofilms when used along with antibiotics.
- Antimicrobial peptides (AMPs) are mostly cationic short peptides that can inhibit ion channels, protein transport and enzymatic activities. They are diverse in their nature, activities, and sources.
- Plant-derived AMPs: Plants produce AMPs as a component of their innate defence mechanism against pathogens. Some of the significant plant-derived AMPs include defensins, 2S albumins, glycine-rich proteins, lipid transfer proteins, snakins, thionins, cyclotides, and napins [60]. For instance, a recombinant plant AMP from tomatoes, when overexpressed in E. coli, was shown to be active against a host of fungi and bacteria [61].
- Insect-derived AMPs: Insect-derived AMPs present excellent alternatives to traditional antibiotics. Four distinct categories of insect AMPs exist: defensins, cecropins, drococins, and attacins. These have been found to be effective against bacteria such as E. coli [62]. Insect-derived AMPs play a significant role in combating many Gram-negative pathogens, for example, cecropins produced by Diptera and Lepidoptera [63].
- Bacterial AMPs (bacteriocins): Bacteriocins are bacterial AMPs. They are highly effective at inhibiting a variety of bacterial species. For example, pyocin, a phage-derived protein synthesized by P. aeruginosa, provides a selective advantage to its host organism within a mixed population [64]. Another bacterial AMP, nisin, which is produced by Lactococcus lactis, has been approved as an alternative to antibiotics for combating Gram-negative pathogens [65].
- Animal AMPs: Tachyplesin III, an AMP derived from the horseshoe crab, has been demonstrated to effectively clear infections of P. aeruginosa and A. baumannii in mouse models [66]. Esculentin-1, an AMP derived from frogs of the genus Rana, was found to be potent against Gram-negative pathogens such as P. aeruginosa and E. coli. Likewise, Brevinin-2Ta displayed potency against Klebsiella [67,68]. Additionally, temporin-1Ja derived from the skin secretions of Rana japonica was able to neutralize endotoxin [69].Recently, mammalian AMPs are found to be very important in combatting antibiotic resistant bacteria. The mammalian AMPs are categorized into two groups namely: Cathelicidins and Defensins. Cathelicidins have shown broad spectrum antimicrobial action. Bovine defensins were shown to be active against various Gram-negative pathogens such as E.coli, Klebsiella pneumoniae and Pseudomonas aeruginosa. Equine α and β Defensins also were observed to show broad spectrum antimicrobial action against Gram-negative pathogens. Furthermore, these peptides can stimulate the immune system to act against evading bacteria. Certain mammals can produce proline rich antimicrobial peptides those show profound antimicrobial activity by inhibiting bacterial protein synthesis. Mammalian milk is an important source of AMPs. Lactalbumin, β-Lactoglobulin, lactoferrin and many others function as potent antibacterial compound. For example, β-Casein 211-225 from human milk was found to show high activity against E.coli and Y. enterocolitica.
- DCAP is a broad-spectrum antibiotic that exhibits potential bactericidal effects against various strains of both Gram-positive and Gram-negative bacteria, including E. coli and P. aeruginosa. As a membrane-targeting drug, DCAP inflicts cellular damage on both Gram-positive and Gram-negative bacteria through 2 primary mechanisms. First, it enhances ion transport across the membrane, which reduces the membrane potential and results in the mislocalization of 2 crucial membrane proteins, MinD and FtsA. Second, it decreases the permeability of the lipid bilayer [70,71]. DCAP is a membrane-active antibiotic that also possesses the added benefit of eliminating dormant bacteria and biofilms.
- Odilorhabdins can effectively kill both Gram-positive and Gram-negative bacteria, particularly ceftazidime-resistant Enterobacteriaceae. They function as inhibitors by binding to specific subunits of bacterial ribosomes and interacting with rRNA and transfer RNA (tRNA). This interaction decodes the translation machinery and enhances the binding affinity of non-cognate aminoacyl tRNAs for the ribosome [3,72].
- Repurposed marketed drugs can provide an effective solution to the issue of antibiotic resistance. Ciclopirox, an antifungal drug, has demonstrated significant antibacterial activity against high-priority, multidrug-resistant Gram-negative bacteria such as A. baumannii, E. coli, and Klebsiella. It has been found to inhibit the synthesis of lipopolysaccharide in Gram-negative bacteria and chelate iron, leading to bacterial inhibition. The anti-diarrheal drug loperamide, when used synergistically with various antibiotics like tetracycline, cephalosporin, and polymyxin B, has been shown to sensitize Gram-negative bacteria to Gram-positive antibiotics such as novobiocin. This may result in changes to the shape of the bacterial cell, leading to dysregulation of the cell’s influx and efflux mechanisms. Berberine, another anti-diarrheal drug, has shown efficacy against multidrug-resistant Mycobacterium tuberculosis and MRSA. A prime example of a repurposed drug is niclosamide, an anti-helminthic drug. Niclosamide has been found to inhibit quorum sensing, leading to the subsequent inhibition of virulence factors and biofilm formation in P. aeruginosa [73]. Clofoctol, a drug used for treating upper respiratory tract and tracheobronchial infections, has also been found to significantly reduce biofilm formation in P. aeruginosa [74].
- Microbial chemical communication, or quorum sensing, refers to the coordinated regulation of gene clusters by a community. This regulation governs processes such as the production of virulence factors, antibiotic sensitivity, and biofilm formation. Autoinducer-2 (AI-2) is a so-called universal autoinducer that controls intraspecies and interspecies bacterial communication [75]. In Gram-positive bacteria, oligopeptides are the most common quorum sensing mediators. However, in Gram-negative bacteria, N-acyl homoserine lactones are the most prevalent. Autoinducer analogues have been demonstrated to be effective quorum sensing inhibitors. For instance, thiolactone analogues of autoinducers have been shown to be potent quorum sensing inhibitors in E. coli, while 3-amino-2-oxazolidinone analogues were found to be effective against P. aeruginosa [76]. Tests have also been conducted on non-analogue quorum sensing inhibitors. For example, halogenated furanones have been reported to reduce quorum sensing in Pseudomonas [76].
- Glycomimetics, which emulate carbohydrate structures, can act as inhibitors of many natural ligands involved in the bacterial pathogenesis pathway. Urinary tract infections caused by uropathogenic E. coli are a major concern, as these bacteria are typically multidrug-resistant and have recently developed resistance to last-resort antibiotics such as colistin and carbapenems. These bacteria produce type 1 fimbriae, which are necessary for their colonization, biofilm formation, and pathogenicity. The FimH adhesin, located at the distal tip of these appendages, binds to mannosylated glycoproteins on urinary tract epithelial cells, promoting bacterial adhesion. Small-molecule mannoside ligand antagonists, a type of glycomimetic, target FimH and subsequently inhibit bacterial colonization [77].
- Short antisense oligonucleotides (ASOs) are currently being explored as a powerful alternative to antibiotics for treating Gram-negative bacteria. These ASOs target the RNA products of specific essential genes within the bacteria. These “programmable RNA antibiotics” are undergoing testing against several different pathogens, including E. coli, Acinetobacter, Campylobacter, Haemophilus, Klebsiella, Pseudomonas, and Salmonella [78,79]. The inherent instability of RNAs is addressed by creating synthetic modified polymers, often attached to a polypeptide backbone. These polymers are also engineered to resist nucleases and proteases. Various types of these modified synthetic ASOs are available, such as locked nucleic acids (LNA), phosphorodiamidate morpholino oligomers (PMO), and peptide nucleic acids (PNA) [80]. However, this method has a significant limitation. To date, it has been targeted towards a limited set of well-defined pathogens, as listed above. Yet, information about the vast majority of microbiota remains unknown. Consequently, ASO-based targeting is challenging for many other human pathogens, such as the human commensal turned pathogen Fusobacterium nucleatum. This bacterium has garnered significant attention due to its association with human diseases such as colorectal cancer, making it a potential target for ASO-based therapy, which could selectively target F. nucleatum in the colon. However, due to the sparse transcriptomic data available for this bacterium, pursuing ASO therapy for this specific purpose is difficult [81].
- Nanoparticles (NPs) which are organic, inorganic, or hybrid particles ≤500 nm in size, are currently under evaluation as an alternative treatment for drug-resistant bacteria. Nanomaterials target a broad spectrum, including the cell wall/cell membrane, reactive oxygen species (ROS) generation, and intracellular component damage. Various types of metal-based and carbon-based NPs, such as quantum dots, polymeric NPs, nanocomposites, and smart nanomaterials, are being utilized against bacteria. These are effective against both planktonic forms and bacterial biofilms. Compared to conventional antibiotics, NPs are less susceptible to existing bacterial resistance mechanisms [82]. Carbon-based NPs have been found to inhibit Klebsiella oxytoca and P. aeruginosa–mediated biofilm formation [83]. Polymeric NPs, such as glycopeptide dendrimers, have also been found to inhibit P. aeruginosa biofilms [84]. Star-shaped polymeric peptide NPs (SNAPPs) have been reported to exert activity against ESKAPE pathogens, which comprise E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter species and are violent nosocomial pathogens [85]. SNAPPs employ a range of bactericidal mechanisms, from membrane disruption and ion exchange to apoptosis-like death mechanisms. Most notably, SNAPPs have been successful in inhibiting colistin-resistant Gram-negative infections and remain potent against multidrug-resistant A. baumannii, which has yet to develop resistance against SNAPPs. Liposome-based NPs have been found to restore the potency of several antibiotics against certain drug-resistant Gram-negative pathogens [86,87].
- Other antibiotic alternatives are currently under study and have shown efficacy.
- Carbon dots are utilized as drugs or drug carriers, demonstrating exceptional efficiency in real-time monitoring of treatment processes. Their antibacterial action is light-driven, and the side effects of their actions are minimal. The mechanism of action involves the photo-induced production of ROS, which damage bacterial DNA, RNA, and proteins. This process also disrupts the bacterial membrane. Broad-spectrum antibacterial action by carbon dots has been observed in cases involving multidrug-resistant E. coli [88]. Given their relative affordability and minimal side effects, these materials could potentially serve as an alternative to antibiotics.
- Silver-doped phosphate coacervates serve as effective alternatives to antibiotics due to their ability to sustainably release silver, which exhibits a significant antimicrobial effect. These coacervates have demonstrated potent activity against wound-related infections, particularly those associated with E. coli and P. aeruginosa [89].
- Royal jelly (RJ) is a yellowish-white viscous fluid formed by worker honeybees. It is known for its ability to inhibit disease-causing bacteria, particularly those associated with periodontal diseases, specifically targeting anaerobic ones [90]. RJ has also been proven to be a potent anti-biofilm agent, capable of inhibiting clinical isolates of P. aeruginosa as well [91]. Another advantage of RJ treatment is its relative lack of side effects.
- Multidrug-resistant Gram-negative pathogens can be targeted by predatory bacteria such as Bdellovibrio spp. and Micavibrio spp. These are Gram-negative bacteria and exhibit a growth phase within the host bacteria. Therefore, they can be used as alternatives to antibiotics and can be used as tools against multidrug-resistant bacteria. Bdellovibrio bacteriovorus HD100 has been reported to reduce the growth of multidrug-resistant Gram-negative bacteria, such as A. baumannii, E. coli, K. pneumoniae, and P. aeruginosa. Micavibrio aeruginosavorus ARL-13 was found to prey on P. aeruginosa and K. pneumoniae.
- Nanobodies are heavy chain-only antibodies of the IgG subtype, possessing a highly stable antigen-binding region. This region allows the nanobodies to bind to and inactivate pathogen proteins, thereby inhibiting the pathogens. A study involving mice demonstrated that nanobodies can protect these animals from infection by enterotoxigenic E. coli [92].
Treatment
β-Lactamase inhibitors
Clavulanic acid and penicillin-based sulfones
Boronic acids as transition state analogues
Efflux pump inhibitors
Membrane permeabilizers
Bacteriophages
Direct-acting compounds
Plant-derived antimicrobial compounds
Antimicrobial peptides
DCAP (2-((3,6-dichloro-9H-carbazol-9-yl)-2-hydroxypropyl)amino)-2-(hydroxymethyl)propane-1,3-diol
Odilorhabdins
Use of repurposed drugs
Anti-virulence therapy
Quorum sensing inhibitors
Fimbriae antagonists (glycomimetic-based therapy)
RNA-based therapeutics
Nanomaterial-based therapy
Miscellaneous
Carbon dots/carbon quantum dots
Silver-doped phosphate coacervates
Royal jelly nutraceutical
Predatory bacteria
Nanobodies
- Multidrug-resistant Gram-negative pathogens, specifically those on the WHO priority bacteria list, are a major focus in public health. Promising new techniques are being developed to overcome the innate and acquired resistance of Gram-negative bacteria. These approaches are characterized by a diverse range of scientific concepts. Therapies such as the use of β-lactamase-inhibiting adjuvants in combination with antibiotics have proven effective in inhibiting the growth of resistant strains of Gram-negative bacteria. Another promising approach is to explore nature for antibacterial agents that can be used as targeted therapeutics, such as bacteriophages, RNA-based therapies, fimbriae antagonists, nanobodies, and AMPs. These methods show promise in combating the silent pandemic of antimicrobial resistance. Indeed, a vast number of alternative approaches are currently under investigation. However, there are several challenges. Translational hurdles are not being adequately addressed. There is a clear trend towards narrow-spectrum or even pathogen-specific approaches, which require a highly developed diagnostic infrastructure that may not be accessible in developing and underdeveloped nations. Adjunctive therapies require an active antibacterial drug and therefore do not fully address the problem of current antibiotic resistance.
- Misuse and overreliance on antibiotics have contributed to the global spread of antibiotic resistance. It is crucial to educate people on the proper use and dispensation of antibiotics, and to implement further regulations and controls. The WHO’s list of priority diseases serves as both an incentive and a framework for developing new antimicrobials and combining new and older medications to combat the rise in multidrug-resistant Gram-negative infections. The success of these novel antibacterial drugs hinges on increased efforts by global governments to stimulate research and development, alongside comprehensive antimicrobial stewardship initiatives and specific local resistance knowledge. To ensure the efficacy of future antimicrobial therapies, protocols must be established to prevent the incorrect prescribing and misuse of antimicrobial drugs in agriculture.
Conclusion
- Antibiotic resistance poses a significant public health concern. This review explores various antibiotic alternatives and auxiliary agents that could potentially address the issue of antibiotic resistance. The primary organisms of focus are Gram-negative pathogens, specifically those on the World Health Organization priority list. The new approaches under consideration are conceptually diverse and innovative, with particular emphasis on nontraditional approaches such as the use of predatory bacteria or engineered phages. While the potential of these innovative ideas is vast, their link to clinical application remains tenuous. Increased global effort, focus, and funding are essential to combat the silent epidemic of antibiotic resistance.
HIGHLIGHTS
-
Ethics Approval
Not applicable.
-
Conflicts of Interest
The authors have no conflicts of interest to declare.
-
Funding
None.
-
Availability of Data
All data generated or analyzed during this study are included in this published article. For other data, these may be requested through the corresponding author.
-
Authors’ Contributions
Conceptualization: PB; Data curation: all authors; Formal analysis: PB, BM; Funding acquisition: NA; Investigation: PB, BM; Methodology: BM, PR, AC; Project administration: PB; Resources: PB; Software: PB; Supervision: PB; Validation: PB, BM; Visualization: BM, AC, PR; Writing–original draft: PB, BM; Writing–review & editing: all authors. All authors read and approved the final manuscript.
Article information
- 1. Domalaon R, Idowu T, Zhanel GG, et al. Antibiotic hybrids: the next generation of agents and adjuvants against Gram-negative pathogens? Clin Microbiol Rev 2018;31:e00077−17.ArticlePubMedPMCPDF
- 2. Chelkeba L, Melaku T, Mega TA. Gram-negative bacteria isolates and their antibiotic-resistance patterns in patients with wound infection in Ethiopia: a systematic review and meta-analysis. Infect Drug Resist 2021;14:277−302.ArticlePubMedPMCPDF
- 3. Breijyeh Z, Jubeh B, Karaman R. Resistance of Gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules 2020;25:1340. ArticlePubMedPMC
- 4. Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018;18:318−27.PubMed
- 5. Kim HS, Kim S, Shin SJ, et al. Gram-negative bacteria and their lipopolysaccharides in Alzheimer’s disease: pathologic roles and therapeutic implications. Transl Neurodegener 2021;10:49. ArticlePubMedPMCPDF
- 6. Ghai I. A Barrier to entry: examining the bacterial outer membrane and antibiotic resistance. Appl Sci 2023;13:4238. Article
- 7. Pages JM, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol 2008;6:893−903.ArticlePubMedPDF
- 8. Masi M, Pages JM. Structure, function and regulation of outer membrane proteins involved in drug transport in Enterobactericeae: the OmpF/C - TolC case. Open Microbiol J 2013;7:22−33.ArticlePubMedPMCPDF
- 9. Huang L, Wu C, Gao H, et al. Bacterial multidrug efflux pumps at the frontline of antimicrobial resistance: an overview. Antibiotics (Basel) 2022;11:520. ArticlePubMedPMC
- 10. Strathdee SA, Davies SC, Marcelin JR. Confronting antimicrobial resistance beyond the COVID-19 pandemic and the 2020 US election. Lancet 2020;396:1050−3.ArticlePubMedPMC
- 11. Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011;17:1791−8.ArticlePubMedPMC
- 12. Isler B, Aslan AT, Akova M, Harris P, Paterson DL. Treatment strategies for OXA-48-like and NDM producing Klebsiella pneumoniae infections. Expert Rev Anti Infect Ther 2022;20:1389−400.ArticlePubMed
- 13. Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect 2006;12:826−36.ArticlePubMed
- 14. Tooke CL, Hinchliffe P, Bragginton EC, et al. β-Lactamases and β-lactamase inhibitors in the 21st century. J Mol Biol 2019;431:3472−500.ArticlePubMedPMC
- 15. Nezhadi J, Narenji H, Soroush Barhaghi MH, et al. Peptide nucleic acid-mediated re-sensitization of colistin resistance Escherichia coli KP81 harboring mcr-1 plasmid. Microb Pathog 2019;135:103646. ArticlePubMed
- 16. Mach T, Neves P, Spiga E, et al. Facilitated permeation of antibiotics across membrane channels: interaction of the quinolone moxifloxacin with the OmpF channel. J Am Chem Soc 2008;130:13301−9.ArticlePubMed
- 17. Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 2009;1794:808−16.ArticlePubMed
- 18. Zander E, Chmielarczyk A, Heczko P, et al. Conversion of OXA-66 into OXA-82 in clinical Acinetobacter baumannii isolates and association with altered carbapenem susceptibility. J Antimicrob Chemother 2013;68:308−11.ArticlePubMed
- 19. Da Silva GJ, Domingues S. Interplay between colistin resistance, virulence and fitness in Acinetobacter baumannii. Antibiotics (Basel) 2017;6:28. ArticlePubMedPMC
- 20. Chung The H, Boinett C, Pham Thanh D, et al. Dissecting the molecular evolution of fluoroquinolone-resistant Shigella sonnei. Nat Commun 2019;10:4828. ArticlePubMedPMCPDF
- 21. Doi Y, de Oliveira Garcia D, Adams J, Paterson DL. Coproduction of novel 16S rRNA methylase RmtD and metallo-beta-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob Agents Chemother 2007;51:852−6.ArticlePubMedPDF
- 22. Yokoyama K, Doi Y, Yamane K, et al. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 2003;362:1888−93.ArticlePubMed
- 23. Eichenberger EM, Thaden JT. Epidemiology and mechanisms of resistance of extensively drug resistant Gram-negative bacteria. Antibiotics (Basel) 2019;8:37. ArticlePubMedPMC
- 24. Doi Y, Adams JM, Yamane K, et al. Identification of 16S rRNA methylase-producing Acinetobacter baumannii clinical strains in North America. Antimicrob Agents Chemother 2007;51:4209−10.ArticlePubMedPMCPDF
- 25. Lee JY, Ko KS. Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn Microbiol Infect Dis 2014;78:271−6.ArticlePubMed
- 26. Albasha AM, Elnosh MM, Osman EH, et al. Helicobacter pylori 23S rRNA gene A2142G, A2143G, T2182C, and C2195T mutations associated with clarithromycin resistance detected in Sudanese patients. BMC Microbiol 2021;21:38. ArticlePubMedPMCPDF
- 27. Webber MA, Bailey AM, Blair JM, et al. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J Bacteriol 2009;191:4276−85.ArticlePubMedPMCPDF
- 28. Ricci V, Tzakas P, Buckley A, et al. Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob Agents Chemother 2006;50:38−42.ArticlePubMedPMCPDF
- 29. McNeil HE, Alav I, Torres RC, et al. Identification of binding residues between periplasmic adapter protein (PAP) and RND efflux pumps explains PAP-pump promiscuity and roles in antimicrobial resistance. PLoS Pathog 2019;15:e1008101.ArticlePubMedPMC
- 30. Laws M, Shaaban A, Rahman KM. Antibiotic resistance breakers: current approaches and future directions. FEMS Microbiol Rev 2019;43:490−516.ArticlePubMedPMCPDF
- 31. Bush K, Bradford PA. Interplay between β-lactamases and new β-lactamase inhibitors. Nat Rev Microbiol 2019;17:295−306.ArticlePubMedPDF
- 32. Salahuddin P, Kumar A, Khan AU. Structure, function of serine and metallo-β-lactamases and their inhibitors. Curr Protein Pept Sci 2018;19:130−44.ArticlePubMed
- 33. Bush K. Past and present perspectives on β-lactamases. Antimicrob Agents Chemother 2018;62:e01076−18.ArticlePubMedPMCPDF
- 34. Yahav D, Giske CG, Gramatniece A, et al. New β-lactam-β-lactamase inhibitor combinations. Clin Microbiol Rev 2020;34:e00115−20.ArticlePDF
- 35. Lizza BD, Betthauser KD, Ritchie DJ, et al. New perspectives on antimicrobial agents: ceftolozane-tazobactam. Antimicrob Agents Chemother 2021;65:e0231820.ArticlePubMedPDF
- 36. Cedano J, Baez M, Pasteran F, et al. Zidebactam restores sulbactam susceptibility against carbapenem-resistant Acinetobacter baumannii isolates. Front Cell Infect Microbiol 2022;12:918868. ArticlePubMedPMC
- 37. Khan Z, Iregui A, Landman D, et al. Activity of cefepime/zidebactam (WCK 5222) against Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter baumannii endemic to New York City medical centres. J Antimicrob Chemother 2019;74:2938−42.ArticlePubMedPDF
- 38. Krajnc A, Lang PA, Panduwawala TD, et al. Will morphing boron-based inhibitors beat the β-lactamases? Curr Opin Chem Biol 2019;50:101−10.ArticlePubMedPMC
- 39. Wenzler E, Scoble PJ. An appraisal of the pharmacokinetic and pharmacodynamic properties of meropenem-vaborbactam. Infect Dis Ther 2020;9:769−84.ArticlePubMedPMCPDF
- 40. Poisson J, Le Hir A, Goutarel R, et al. Isolation of reserpine from roots of Rauwolfia vomitoria Afz. C R Hebd Seances Acad Sci 1954;238:1607−9.PubMed
- 41. Zhou XY, Ye XG, He LT, et al. In vitro characterization and inhibition of the interaction between ciprofloxacin and berberine against multidrug-resistant Klebsiella pneumoniae. J Antibiot (Tokyo) 2016;69:741−6.ArticlePubMedPMCPDF
- 42. Gbian DL, Omri A. The impact of an efflux pump inhibitor on the activity of free and liposomal antibiotics against Pseudomonas aeruginosa. Pharmaceutics 2021;13:577. ArticlePubMedPMC
- 43. Reza A, Sutton JM, Rahman KM. Effectiveness of efflux pump inhibitors as biofilm disruptors and resistance breakers in Gram-negative (ESKAPEE) bacteria. Antibiotics (Basel) 2019;8:229. ArticlePubMedPMC
- 44. Mahamoud A, Chevalier J, Baitiche M, et al. An alkylaminoquinazoline restores antibiotic activity in Gram-negative resistant isolates. Microbiology (Reading) 2011;157(Pt 2). 566−71.ArticlePubMed
- 45. Farrag HA, Abdallah N, Shehata MM, et al. Natural outer membrane permeabilizers boost antibiotic action against irradiated resistant bacteria. J Biomed Sci 2019;26:69. ArticlePubMedPMCPDF
- 46. Song J, Xia F, Jiang H, et al. Identification and characterization of HolGH15: the holin of Staphylococcus aureus bacteriophage GH15. J Gen Virol 2016;97:1272−81.ArticlePubMed
- 47. Bai J, Lee S, Ryu S. Identification and in vitro characterization of a novel phage endolysin that targets Gram-negative bacteria. Microorganisms 2020;8:447. ArticlePubMedPMC
- 48. Rahman MU, Wang W, Sun Q, et al. Endolysin, a promising solution against antimicrobial resistance. Antibiotics (Basel) 2021;10:1277. ArticlePubMedPMC
- 49. Kim S, Jin JS, Choi YJ, et al. LysSAP26, a new recombinant phage endolysin with a broad spectrum antibacterial activity. Viruses 2020;12:1340. ArticlePubMedPMC
- 50. Fursov MV, Abdrakhmanova RO, Antonova NP, et al. Antibiofilm activity of a broad-range recombinant endolysin LysECD7: in vitro and in vivo study. Viruses 2020;12:545. ArticlePubMedPMC
- 51. Zampara A, Sorensen MC, Grimon D, et al. Exploiting phage receptor binding proteins to enable endolysins to kill Gram-negative bacteria. Sci Rep 2020;10:12087. ArticlePubMedPMCPDF
- 52. Merabishvili M, Pirnay JP, De Vos D. Guidelines to compose an ideal bacteriophage cocktail. Methods Mol Biol 2018;1693:99−110.ArticlePubMed
- 53. Jault P, Leclerc T, Jennes S, et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect Dis 2019;19:35−45.ArticlePubMed
- 54. Huang G, Wei Z, Wang D. What do we lean from the “PhagoBurn” project. Burns 2019;45:260. ArticlePubMed
- 55. Shriram V, Khare T, Bhagwat R, et al. Inhibiting bacterial drug efflux pumps via phyto-therapeutics to combat threatening antimicrobial resistance. Front Microbiol 2018;9:2990. ArticlePubMedPMC
- 56. Anand U, Jacobo-Herrera N, Altemimi A, et al. A comprehensive review on medicinal plants as antimicrobial therapeutics: potential avenues of biocompatible drug discovery. Metabolites 2019;9:258. ArticlePubMedPMC
- 57. Khare T, Anand U, Dey A, et al. Exploring phytochemicals for combating antibiotic resistance in microbial pathogens. Front Pharmacol 2021;12:720726. ArticlePubMedPMC
- 58. Dahiya P, Purkayastha S. Phytochemical screening and antimicrobial activity of some medicinal plants against multi-drug resistant bacteria from clinical isolates. Indian J Pharm Sci 2012;74:443−50.ArticlePubMedPMC
- 59. Owen L, Laird K. Synchronous application of antibiotics and essential oils: dual mechanisms of action as a potential solution to antibiotic resistance. Crit Rev Microbiol 2018;44:414−35.ArticlePubMed
- 60. Baindara P, Mandal SM. Plant-derived antimicrobial peptides: novel preservatives for the food industry. Foods 2022;11:2415. ArticlePubMedPMC
- 61. Herbel V, Schafer H, Wink M. Recombinant production of Snakin-2 (an antimicrobial peptide from tomato) in E. coli and analysis of its bioactivity. Molecules 2015;20:14889−901.ArticlePubMedPMC
- 62. Di Somma A, Moretta A, Cane C, et al. Structural and functional characterization of a novel recombinant antimicrobial peptide from Hermetia illucens. Curr Issues Mol Biol 2021;44:1−13.ArticlePubMedPMC
- 63. Wu Q, Patocka J, Kuca K. Insect antimicrobial peptides, a mini review. Toxins (Basel) 2018;10:461. ArticlePubMedPMC
- 64. Kim BO, Kim ES, Yoo YJ, et al. Phage-derived antibacterials: harnessing the simplicity, plasticity, and diversity of phages. Viruses 2019;11:268. ArticlePubMedPMC
- 65. Dijksteel GS, Ulrich MM, Middelkoop E, et al. Review: lessons learned from clinical trials using antimicrobial peptides (AMPs). Front Microbiol 2021;12:616979. ArticlePubMedPMC
- 66. Qi J, Gao R, Liu C, et al. Potential role of the antimicrobial peptide Tachyplesin III against multidrug-resistant P. aeruginosa and A. baumannii coinfection in an animal model. Infect Drug Resist 2019;12:2865−74.PubMedPMC
- 67. Patocka J, Nepovimova E, Klimova B, et al. Antimicrobial peptides: amphibian host defense peptides. Curr Med Chem 2019;26:5924−46.ArticlePubMed
- 68. Liu S, Long Q, Xu Y, et al. Assessment of antimicrobial and wound healing effects of Brevinin-2Ta against the bacterium Klebsiella pneumoniae in dermally-wounded rats. Oncotarget 2017;8:111369−85.ArticlePubMedPMC
- 69. Rosenfeld Y, Barra D, Simmaco M, et al. A synergism between temporins toward Gram-negative bacteria overcomes resistance imposed by the lipopolysaccharide protective layer. J Biol Chem 2006;281:28565−74.ArticlePubMed
- 70. Eun YJ, Foss MH, Kiekebusch D, et al. DCAP: a broad-spectrum antibiotic that targets the cytoplasmic membrane of bacteria. J Am Chem Soc 2012;134:11322−5.ArticlePubMedPMC
- 71. Allavena G, Debellis D, Marotta R, et al. A broad-spectrum antibiotic, DCAP, reduces uropathogenic Escherichia coli infection and enhances vorinostat anticancer activity by modulating autophagy. Cell Death Dis 2018;9:780. ArticlePubMedPMCPDF
- 72. Pantel L, Florin T, Dobosz-Bartoszek M, et al. Odilorhabdins, antibacterial agents that cause miscoding by binding at a new ribosomal site. Mol Cell 2018;70:83−94.ArticlePubMed
- 73. Imperi F, Massai F, Ramachandran Pillai C, et al. New life for an old drug: the anthelmintic drug niclosamide inhibits Pseudomonas aeruginosa quorum sensing. Antimicrob Agents Chemother 2013;57:996−1005.ArticlePubMedPMCPDF
- 74. D’Angelo F, Baldelli V, Halliday N, et al. Identification of FDA-approved drugs as antivirulence agents targeting the pqs quorum-sensing system of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2018;62:e01296−18.ArticlePubMedPMCPDF
- 75. Zhang L, Li S, Liu X, et al. Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat Commun 2020;11:5371. ArticlePubMedPMCPDF
- 76. Duplantier M, Lohou E, Sonnet P. Quorum sensing inhibitors to quench P. aeruginosa pathogenicity. Pharmaceuticals (Basel) 2021;14:1262. ArticlePubMedPMC
- 77. Mydock-McGrane LK, Hannan TJ, Janetka JW. Rational design strategies for FimH antagonists: new drugs on the horizon for urinary tract infection and Crohn’s disease. Expert Opin Drug Discov 2017;12:711−31.ArticlePubMedPMC
- 78. Sully EK, Geller BL. Antisense antimicrobial therapeutics. Curr Opin Microbiol 2016;33:47−55.ArticlePubMedPMC
- 79. Geller BL, Li L, Martinez F, et al. Morpholino oligomers tested in vitro, in biofilm and in vivo against multidrug-resistant Klebsiella pneumoniae. J Antimicrob Chemother 2018;73:1611−9.ArticlePubMedPMC
- 80. Dhuri K, Bechtold C, Quijano E, et al. Antisense oligonucleotides: an emerging area in drug discovery and development. J Clin Med 2020;9:2004. ArticlePubMedPMC
- 81. Vogel J. An RNA biology perspective on species-specific programmable RNA antibiotics. Mol Microbiol 2020;113:550−9.ArticlePubMedPDF
- 82. Pelgrift RY, Friedman AJ. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv Drug Deliv Rev 2013;65:1803−15.ArticlePubMed
- 83. Malek I, Schaber CF, Heinlein T, et al. Vertically aligned multi walled carbon nanotubes prevent biofilm formation of medically relevant bacteria. J Mater Chem B 2016;4:5228−35.ArticlePubMed
- 84. Reymond JL, Bergmann M, Darbre T. Glycopeptide dendrimers as Pseudomonas aeruginosa biofilm inhibitors. Chem Soc Rev 2013;42:4814−22.ArticlePubMed
- 85. Chen YF, Lai YD, Chang CH, et al. Star-shaped polypeptides exhibit potent antibacterial activities. Nanoscale 2019;11:11696−708.ArticlePubMed
- 86. Pushparaj Selvadoss P, Nellore J, Balaraman Ravindrran M, et al. Novel pyochelin-based PEGylated liposomes for enhanced delivery of antibiotics against resistant clinical isolates of Pseudomonas aeruginosa. Artif Cells Nanomed Biotechnol 2018;46:2043−53.PubMed
- 87. Singla S, Harjai K, Katare OP, et al. Encapsulation of bacteriophage in liposome accentuates its entry in to macrophage and shields it from neutralizing antibodies. PLoS One 2016;11:e0153777.ArticlePubMedPMC
- 88. Dong X, Liang W, Meziani MJ, et al. Carbon dots as potent antimicrobial agents. Theranostics 2020;10:671−86.ArticlePubMedPMC
- 89. Nikolaou A, Felipe-Sotelo M, Dorey R, et al. Silver-doped phosphate coacervates to inhibit pathogenic bacteria associated with wound infections: an in vitro study. Sci Rep 2022;12:10778. ArticlePubMedPMCPDF
- 90. Khosla A, Gupta SJ, Jain A, et al. Evaluation and comparison of the antimicrobial activity of royal jelly: a holistic healer against periodontopathic bacteria: an in vitro study. J Indian Soc Periodontol 2020;24:221−6.ArticlePubMedPMC
- 91. Bagameri L, Baci GM, Dezmirean DS. Royal jelly as a nutraceutical natural product with a focus on its antibacterial activity. Pharmaceutics 2022;14:1142. ArticlePubMedPMC
- 92. Amcheslavsky A, Wallace AL, Ejemel M, et al. Anti-CfaE nanobodies provide broad cross-protection against major pathogenic enterotoxigenic Escherichia coli strains, with implications for vaccine design. Sci Rep 2021;11:2751. ArticlePubMedPMCPDF
References
Figure & Data
References
Citations

- Efficacy of new generation biosorbents for the sustainable treatment of antibiotic residues and antibiotic resistance genes from polluted waste effluent
Barkha Madhogaria, Sangeeta Banerjee, Atreyee Kundu, Prasanta Dhak
Infectious Medicine.2024; 3(1): 100092. CrossRef - Evaluation of Plant-Based Silver Nanoparticles for Antioxidant Activity and Promising Wound-Healing Applications
Maria Qubtia, Shazia Akram Ghumman, Sobia Noreen, Huma Hameed, Shazia Noureen, Rizwana Kausar, Ali Irfan, Pervaiz Akhtar Shah, Hafsa Afzal, Misbah Hameed, Mohammad Raish, Maria Rana, Ajaz Ahmad, Katarzyna Kotwica-Mojzych, Yousef A. Bin Jardan
ACS Omega.2024; 9(10): 12146. CrossRef



 Cite
Cite




