Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 14(5); 2023 > Article
-
Original Article
Drug resistance and the genotypic characteristics of rpoB and katG in rifampicin- and/or isoniazid-resistant Mycobacterium tuberculosis isolates in central Vietnam -
Thi Binh Nguyen Nguyen1
 , Thi Kieu Diem Nguyen2
, Thi Kieu Diem Nguyen2 , Van Hue Trương3
, Van Hue Trương3 , Thi Tuyet Ngoc Tran4
, Thi Tuyet Ngoc Tran4 , van Bao Thang Phan4
, van Bao Thang Phan4 , Thi Tuyen Nguyen4
, Thi Tuyen Nguyen4 , Hoang Bach Nguyen4
, Hoang Bach Nguyen4 , Viet Quynh Tram Ngo4
, Viet Quynh Tram Ngo4 , Van Tuan Mai5
, Van Tuan Mai5 , Paola Molicotti6
, Paola Molicotti6
-
Osong Public Health and Research Perspectives 2023;14(5):347-355.
DOI: https://doi.org/10.24171/j.phrp.2023.0124
Published online: October 18, 2023
1Department of Infectious Diseases and Tuberculosis, Hue University of Medicine and Pharmacy, Hue University, Hue, Vietnam
2Da Nang Lung Hospital, Da Nang, Vietnam
3Central Hospital 71, Thanh Hoa, Vietnam
4Department of Microbiology, Hue University of Medicine and Pharmacy, Hue University, Hue, Vietnam
5Department of Microbiology, Hue Central Hospital, Hue, Vietnam
6Department of Biomedical Science, Microbiology and Clinical Microbiology, University of Sassari, Sassari, Italy
- Corresponding author: Thi Binh Nguyen Nguyen Department of Infectious Diseases and Tuberculosis, Hue University of Medicine and Pharmacy, Hue University, Hue, Vietnam E-mail: ntbnguyen@hueuni.edu.vn
© 2023 Korea Disease Control and Prevention Agency.
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
- 1,145 Views
- 76 Download
Abstract
-
Objectives
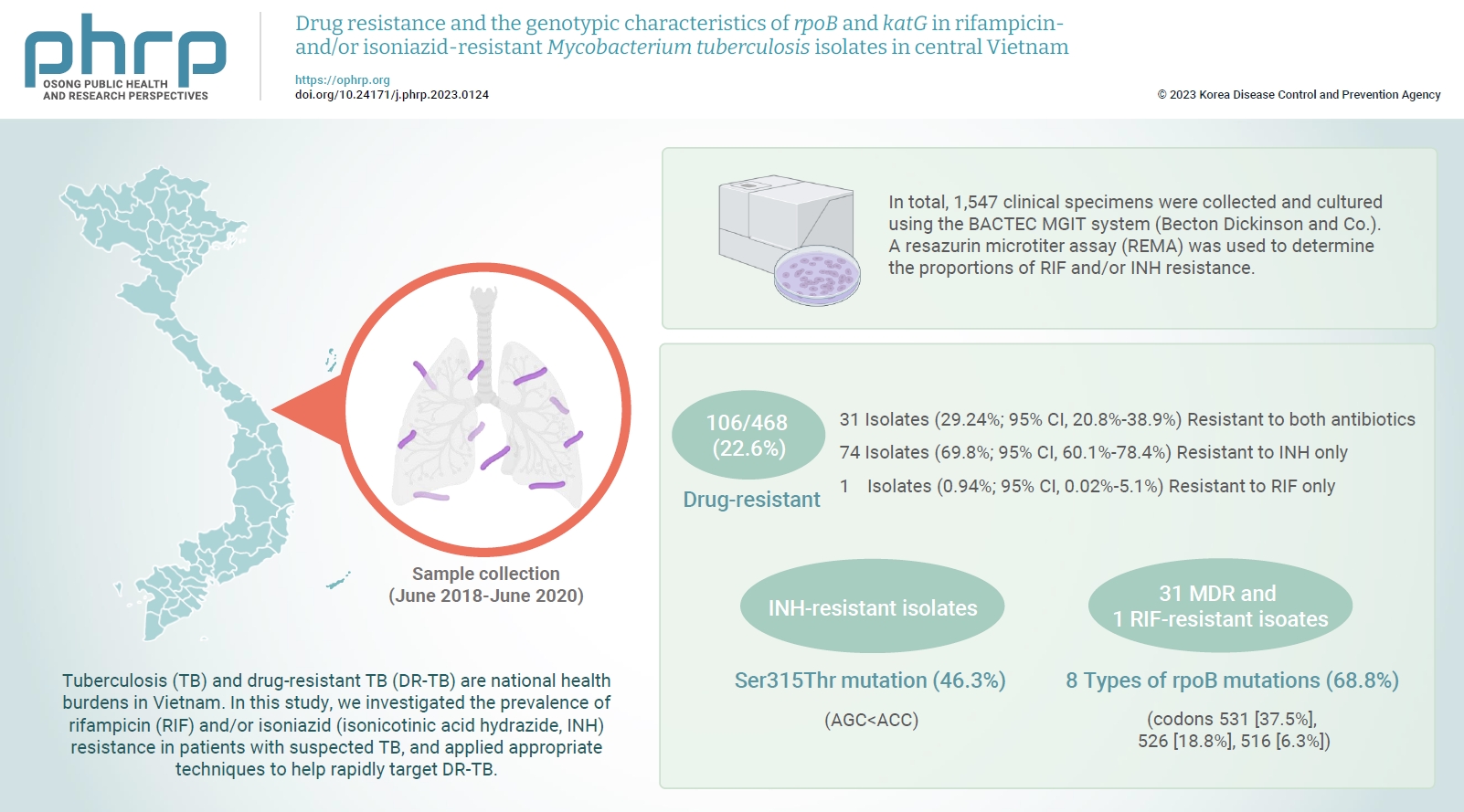
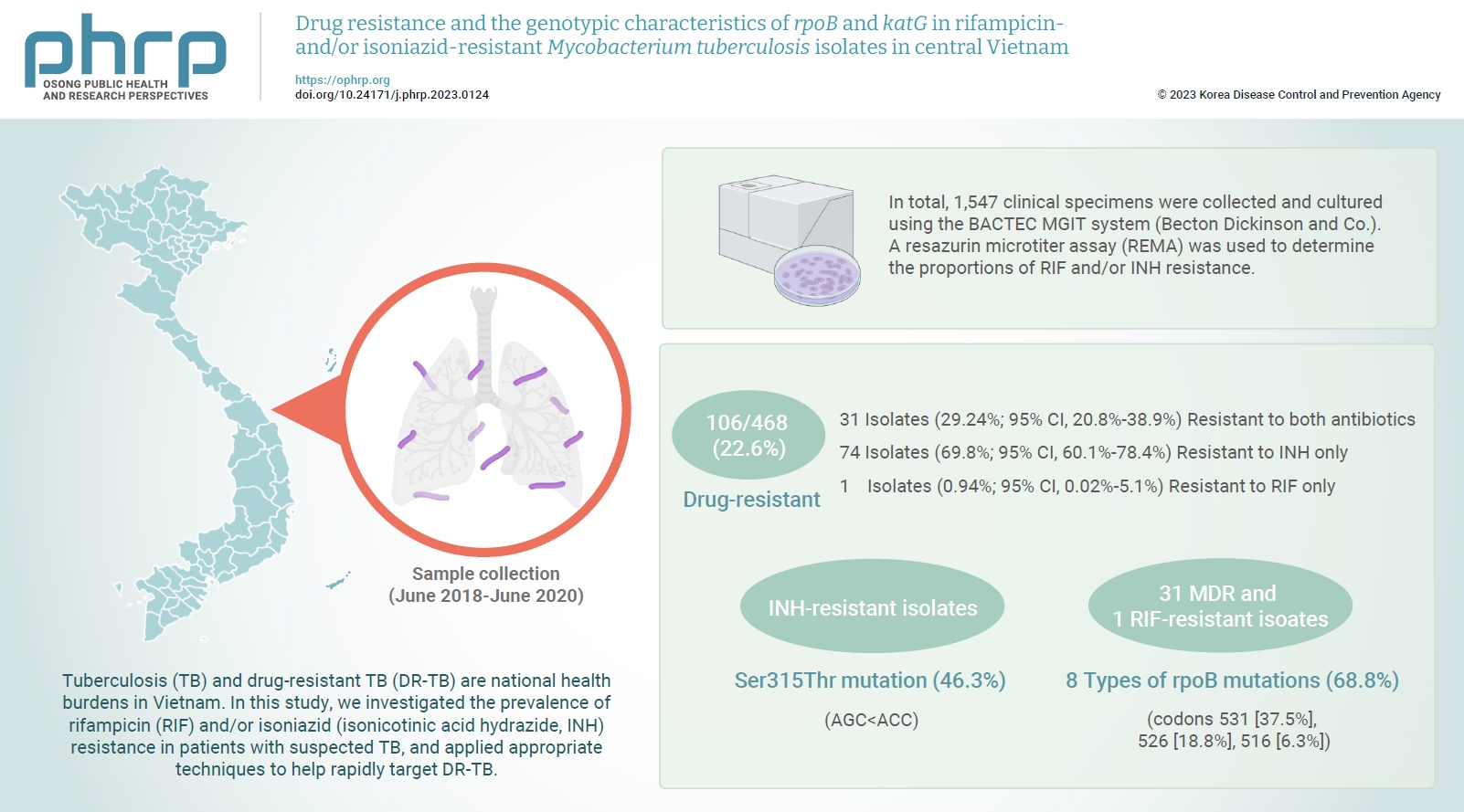
- Tuberculosis (TB) and drug-resistant TB (DR-TB) are national health burdens in Vietnam. In this study, we investigated the prevalence of rifampicin (RIF) and/or isoniazid (isonicotinic acid hydrazide, INH) resistance in patients with suspected TB, and applied appropriate techniques to help rapidly target DR-TB.
-
Methods
- In total, 1,547 clinical specimens were collected and cultured using the BACTEC MGIT system (Becton Dickinson and Co.). A resazurin microtiter assay (REMA) was used to determine the proportions of RIF and/or INH resistance. A real-time polymerase chain reaction panel with TaqMan probes was employed to identify the mutations of rpoB and katG associated with DR-TB in clinical isolates. Genotyping of the identified mutations was also performed.
-
Results
- A total of 468 Mycobacterium tuberculosis isolates were identified using the REMA. Of these isolates, 106 (22.6%) were found to be resistant to 1 or both antibiotics. Of the resistant isolates, 74 isolates (69.8%) were resistant to isoniazid (INH) only, while 1 isolate (0.94%) was resistant to RIF only. Notably, 31 isolates (29.2%) were resistant to both antibiotics. Of the 41 phenotypically INH-resistant isolates, 19 (46.3%) had the Ser315Thr mutation. There were 8 different rpoB mutations in 22 (68.8%) of the RIF-resistant isolates. The most frequently detected mutations were at codons 531 (37.5%), 526 (18.8%), and 516 (6.3%).
-
Conclusion
- To help prevent new cases of DR-TB in Vietnam, it is crucial to gain a comprehensive understanding of the genotypic DR-TB isolates.
- Tuberculosis (TB) poses a significant health burden worldwide, and drug-resistant TB (DR-TB) is one of the biggest challenges to TB treatment [1−3]. Therefore, developing diagnostic methodologies for TB and DR-TB is a critical aspect of TB management. In 2019, the World Health Organization (WHO) estimated that 10.4 million patients had TB, and 1.5 million deaths were attributed to the disease. Vietnam is currently ranked 16th among the 30 countries with high burdens of TB and 13th among the 30 countries with the highest prevalence of DR-TB [4]. Although the Vietnam National Tuberculosis Control Program has nationwide coverage, diagnosing, treating, and managing TB and DR-TB remains a significant public health challenge [5,6]. Commercial drug susceptibility testing (DST) systems such as the liquid-medium BACTEC MGIT 960 (Becton Dickinson and Co.) and molecular DST methods such as the GeneXpert MTB/RIF (Cepheid) provide rapid results and reduce turnaround time [1,7]. However, they require costly reagents, modern equipment, and only detect some drugs, limiting their availability in Vietnamese laboratories. Therefore, developing phenotypic DST for Mycobacterium tuberculosis that determines the minimum inhibitory concentration (MIC) values of many TB drugs will help doctors make better treatment decisions by considering the level of resistance of each drug. These methods are simple, reliable, and cost-effective, making them necessary in countries with a high prevalence of DR-TB [8−10]. Moreover, there are limited genetic studies that characterize the genotype of M. tuberculosis isolates in central Vietnam. A thorough understanding of the genotypic DR-TB isolates helps support a focus on infection control and surveillance to prevent new cases of DR-TB in this region. To reduce the morbidity and mortality of DR-TB, accurate and rapid diagnosis in the early stages of TB is important. This requires implementing a molecular assay that can detect the wide variety of drug-resistant mutations in clinical isolates of M. tuberculosis. Evaluating the molecular assays that can detect a wide variety of M. tuberculosis-resistant mutations in clinical isolates is vital in the effort to reduce DR-TB morbidity and mortality by enabling accurate and rapid diagnosis in the early stages [11,12]. Therefore, to assist in decreasing DR-TB cases in Vietnam, it is necessary to combine phenotypic and molecular methods until more is understood about the clinical relevance of phenotypic susceptibility in isolates with drug-resistant mutation mechanisms. This study evaluated the frequency of TB resistance to rifampicin (RIF) and isoniazid (isonicotinic acid hydrazide, INH), which are the most effective first-line drugs in TB treatment.
Introduction
- Sample Collection
- From June 2019 to June 2020, samples were collected from patients suspected of TB, and M. tuberculosis isolates were cultured for DST at various locations in central Vietnam, including Da Nang Lung Hospital, Central Hospital 71, Thanh Hoa, and the Department of Microbiology at Hue Central Hospital in Hue city. The Carlo Urbani Center’s Department of Microbiology at Hue University of Medicine and Pharmacy conducted the real-time (RT)-polymerase chain reaction (PCR) TaqMan allelic discrimination assays.
- Drug Susceptibility Testing
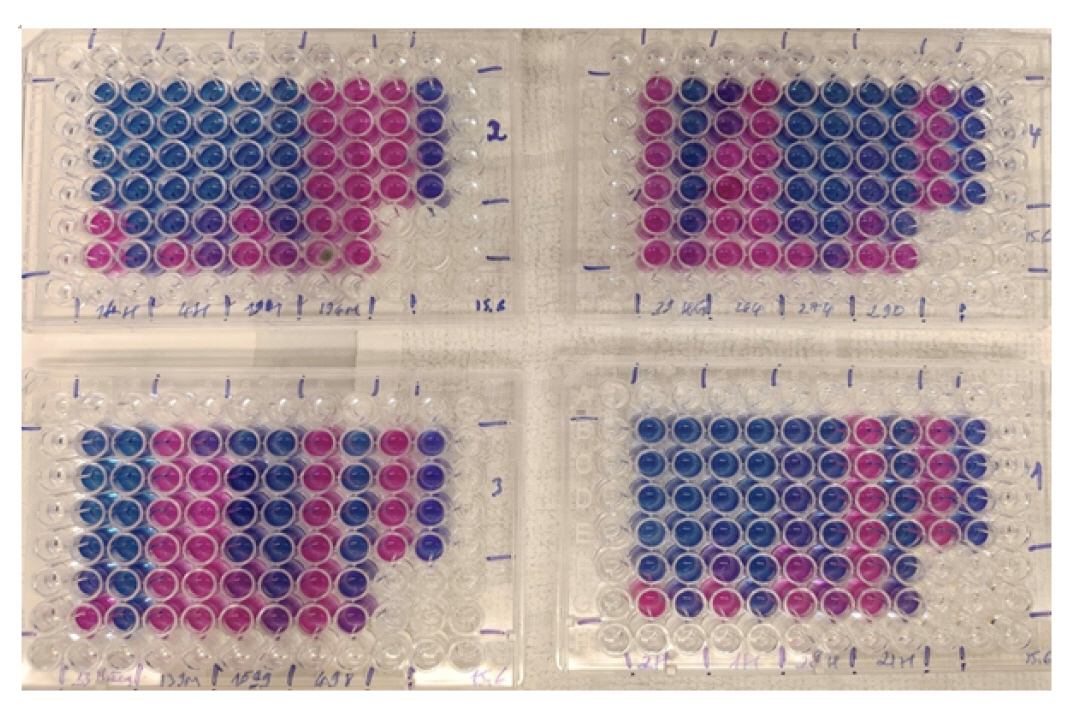
- DST was performed using the resazurin microtiter assay (REMA) method. A total of 1,547 clinical samples were inoculated in the BACTEC MGIT 960 (Becton Dickinson and Co.), of which 500 were found to be positive for M. tuberculosis. The M. tuberculosis strain H37Rv (ATCC 27294) was utilized as the susceptible control strain, while a previously identified multidrug-resistant (MDR)-TB strain from the microbiology department at Da Nang Lung Hospital was employed as the resistant control strain.
- Middlebrook 7H9 broth was dispensed into each well of a Corning 96-well plate for the REMA. All drugs (HiMedia Laboratories Private Ltd.) were used in lyophilized form and subsequently rehydrated and filtered to make the stock solution. INH was diluted with sterile water to reach a concentration of 1 mg/mL, while RIF was diluted with methanol to achieve a concentration of 10 mg/mL. The INH concentration ranged from 1.00 to 0.031 µg/mL, and the RIF concentration ranged from 2.00 to 0.061 µg/mL. To determine the resistance or susceptibility of M. tuberculosis, a strain was considered resistant to INH if the MIC was ≥0.25 µg/mL, while a strain was deemed resistant to RIF if the MIC was ≥0.5 µg/mL. This method, developed by Palomino et al. [9] in 2002 and Martin et al. [13] in 2005 using the colorimetric indicator resazurin, has been proposed for the DST of M. tuberculosis. To confirm continued susceptibility or resistance, independent of the MIC, the results of the REMA were compared with those of the reference method using a liquid culture in the BACTEC MGIT mycobacterial detection system (Becton Dickinson and Co.). The commercial kits that we utilized were supplied with fixed concentrations of 0.1 µg/mL and 1.0 µg/mL, for INH and RIF, respectively (Figure 1) [9,13].
- RT-PCR TaqMan Allelic Discrimination Assay
- A total of 52 clinical isolates of M. tuberculosis (31 MDR, 1 RIF mono-resistant, 10 INH mono-resistant, and 10 drug-sensitive isolates) were used to extract genomic DNA. A 200-µL sample was placed in an Eppendorf tube and 400 µL of InstaGene Matrix (Bio-Rad Laboratories) was added. This was followed by vortexing and incubating the mixture at 100°C for 10 minutes. The mixture was then centrifuged at 14,000 rpm for 2 minutes, and the DNA in the supernatant was collected and stored at −20°C.
- To conduct the allelic discrimination test, primers were designed to amplify a sequence of 208 base pairs (bp) in the rpoB gene and 110 bp in the katG gene. Four TaqMan probes, which could distinguish one-base mismatches, were used to detect genetic variation in the wild-type M. tuberculosis strain (H37Rv) in both rpoB and katG. All primers and probes used in this study were manufactured by Integrated DNA Technologies Inc. (Table 1) [14,15].
- To detect mutations in the selected regions of the rpoB and katG genes, 2 separate reactions were conducted in tube A (for rpoB mutation detection) and tube B (for katG mutation detection) with a final reaction volume of 25 µL. The master mix used in this study was produced by Integrated DNA Technologies Inc. An optimized multiplex-probe RT-PCR reaction was established, and PCR amplification was performed as follows: initial denaturation at 94°C for 10 minutes, followed by 40 cycles at 94°C for 25 seconds and 60°C for 55 seconds, using the Mx3000P qPCR System (Agilent Technologies Inc.).
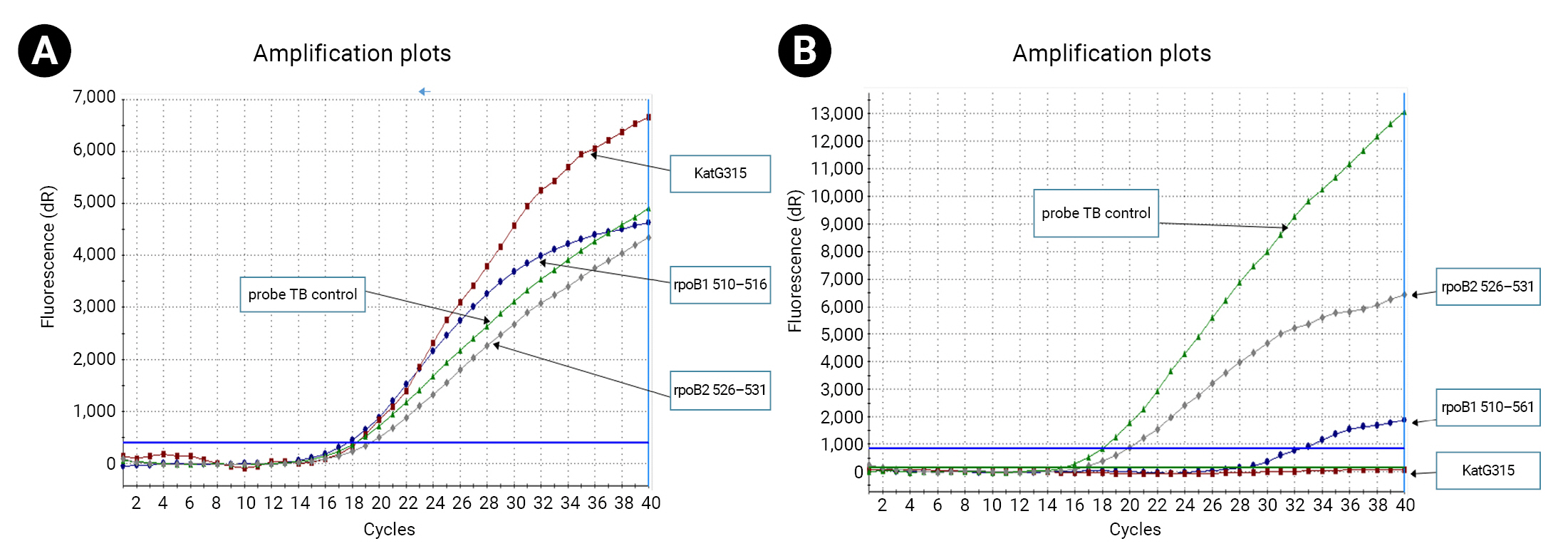
- We determined the cycle threshold (Ct) value from the TB control probe that bound to the 81 bp hot spots outside the target regions in the rpoB gene, and calculated the ∆Ct value, which represented the difference between the control TB probe and each specific probe (∆Ct=mutant Ct−control TB Ct). In the mutant genotypes, a single base variation in the target sequence of rpoB or katG could lead to mismatching of the corresponding probe and failure to combine with the target, resulting in probe dropout from the sequence. This would generate negative fluorescence signals during amplification, and the Ct values of the mutant sequences would be recorded as zero (Ct=0). Ultimately, M. tuberculosis was identified using the TB control probe.
- DNA samples extracted from the 52 isolates that had been identified as M. tuberculosis by RT-PCR, were used to amplify rpoB and katG using the primer sequences presented in Table 1. PCR amplification was performed as follows: an initial denaturation step at 95°C for 10 minutes, followed by 36 cycles of 95°C for 30 seconds, 55°C for 30 seconds, 72°C for 30 seconds, and a final extension step at 72°C for 5 minutes in the Veriti Thermal Cycler (Thermo Fisher Scientific Inc.). The PCR products were then sent to Apical Scientific Sdn Bhd for Sanger sequencing. The sequencing data were initially analyzed using Sequencing Analysis Software ver. 6.0, and quality control was checked by Sequence Scanner Software (both from Thermo Fisher Scientific Inc.).
- Ethics Approval
- Ethical approval for this study was granted by The Ethical Committee in Biomedical Research of the University of Medicine and Pharmacy-Hue University, (no: H2019/350).
- The director of each hospital granted permission to conduct the study. Before specimen collection, patients/guardians were provided with information about the study and asked to provide written informed consent. Participants were informed of their right to voluntarily participate in the study.
Materials and Methods
DNA extraction
Primers and probes
Procedure
Allelic discrimination data analysis
rpoB and katG sequencing
- Among the 1,547 clinical specimens collected from patients suspected of having TB, 32.3% (500/1,547 samples) were identified as positive through the BACTEC MGIT 960 system (Becton Dickinson and Co.). RT-PCR was also able to detect all M. tuberculosis isolates. Of the 500 isolates, 6.4% (32/500 samples) were excluded from the DST because they were heavily contaminated.
- Phenotypic DST by REMA
- A total of 468 M. tuberculosis isolates underwent REMA testing, which revealed that 106 (22.6%) were drug-resistant. Specifically, 74 (69.8%; 95% confidence interval [CI], 60.1%−78.4%) were resistant to INH, while 1 (0.94%; 95% CI, 0.02%−5.1%) was resistant to RIF, and 31 (29.24%; 95% CI, 20.8%−38.9%) were resistant to both antibiotics (MDR-TB strains). When the REMA results were compared to those independently obtained using the BACTEC MGIT 960 system (Becton Dickinson and Co.), the sensitivity and specificity for INH resistance were 100% and 99.19%, respectively. The sensitivity and specificity for RIF resistance were both 100%, the overall concordance for MDR-TB was 99.78%, and the positive predictive value and negative predictive value for MDR-TB detection were 96.77% and 100%, respectively (Table 2).
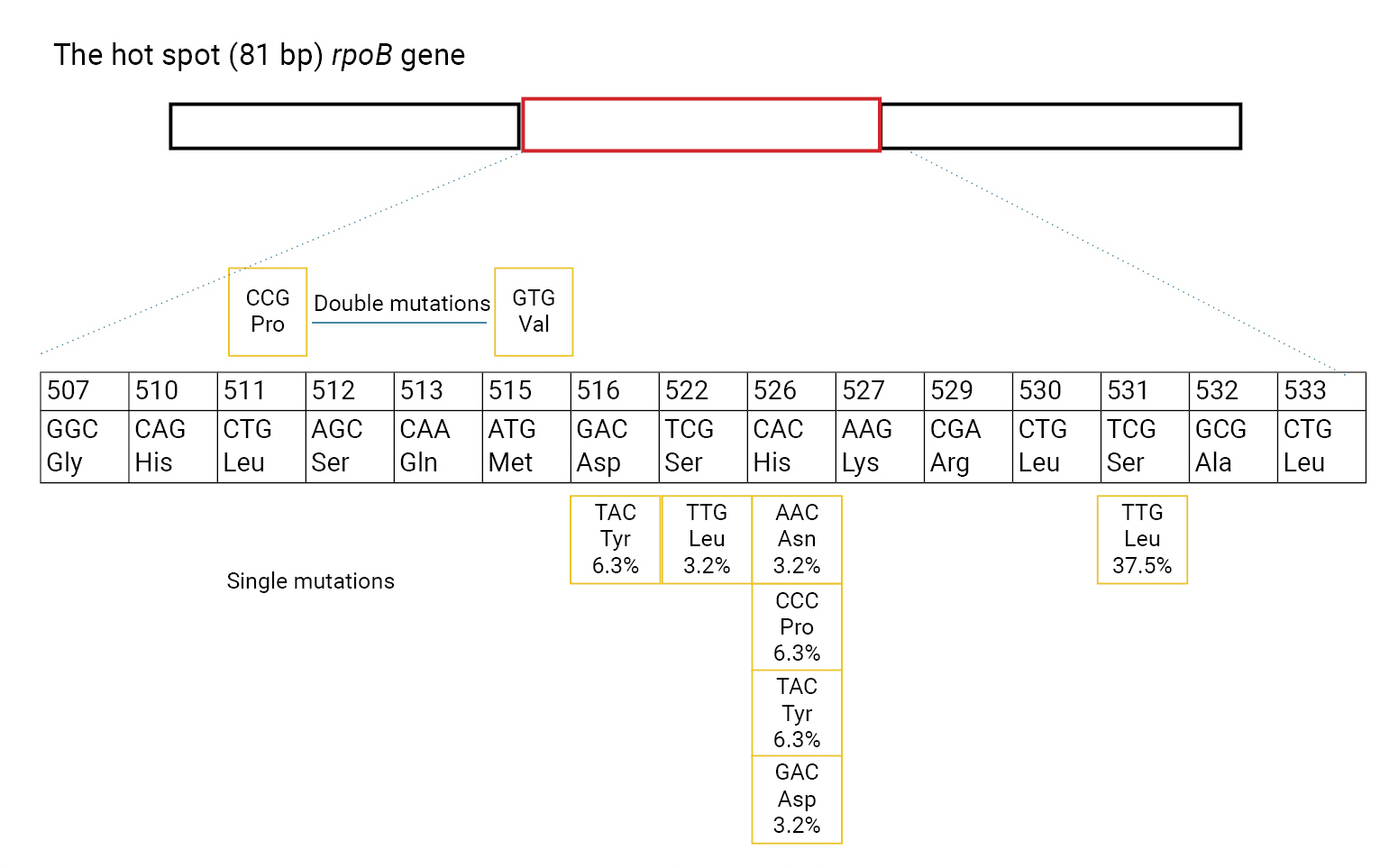
- Genotypic Detection of Drug-Resistant Isolates
- Of the phenotypic INH-resistant isolates, 46.3% (19/41) had a Ser315Thr mutation (AGC>ACC). Among the 31 MDR and 1 RIF-resistant isolates, 22/32 (68.8%) had 8 types of rpoB mutations. The most frequent rpoB mutations were found at codons 531, 526, and 516, accounting for 37.5% (12/32), 18.8% (6/32), and 6.3% (2/32), respectively. Sequencing analysis detected the point mutation Ser522Leu (TCG>TTG) outside of the 2 probes used to determine mutations in rpoB. None of the 11/11 INH-susceptible isolates and 20/20 RIF-susceptible isolates had mutations in katG and rpoB (Figure 2).
- The RT-PCR allelic and DNA sequencing results showed a sensitivity and specificity of 95.5% and 100%, respectively, for detecting INH resistance through mutation analysis of katG codons 311−316. Compared to the sequencing for katG, this method had an accuracy of 98.08%. For the detection of RIF resistance, the RT-PCR allelic and DNA sequencing results had a sensitivity and specificity of 95.00% and 100%, respectively, for mutation analysis in the rpoB codons 510−531 (Figure 3).
Results
- This study evaluated the rate of phenotypic INH and/or RIF resistance in M. tuberculosis isolates from 3 hospitals in central Vietnam using REMA as a DST method. The REMA plate method, which was developed by Palomino et al. [9] in 2002 and Martin et al. [16] in 2003 and uses the colorimetric indicator resazurin, was employed for this purpose. REMA is a useful tool for the DST of both first-line and second-line TB drugs and can also determine resistance levels by assessing the MIC value, thus assisting clinicians in making better treatment decisions.
- In this study, REMA was used to test the drug susceptibility of 500 M. tuberculosis isolates. Although 468 isolates gave interpretable results, 32 strains (6.4%) provided invalid results due to suspected contamination, likely resulting from poor storage. This outcome is consistent with the WHO’s 2020 epidemiological report on DR-TB in Vietnam, which reported a rate of 3.6% for new MDR-TB cases and 17% for previously treated cases. INH resistance can be a precursor to the emergence of MDR-TB, and the drug plays a crucial role in treating TB and latent TB infection [17,18]. However, despite the effectiveness of the GeneXpert MTB/RIF test in detecting M. tuberculosis and RIF resistance in the process of identifying MDR-TB cases, many pulmonary hospital laboratories in Vietnam may miss cases of INH mono-resistance, which are difficult to treat in patients with TB and latent TB infection. Our study suggests that this strategy may not be adequate since a high proportion (99%) of RIF-resistant isolates are also resistant to INH. In addition, REMA is a cost-effective alternative to commercial phenotypic DST and cultures, with an estimated cost of 3 United States dollars (USD) per strain tested, whereas the cultures and DST of many drugs in Vietnam by the BACTEC MGIT 960 can cost up to 35 USD for each strain tested. Therefore, REMA can be an effective method for detecting MDR-TB cases, especially in countries with a high prevalence of TB and limited resources. Its low cost, simplicity, and ease of execution do not demand highly technical skills [16,19]. Finally, the prompt screening of drug resistance in pulmonary hospital laboratories and the quick availability of results will facilitate the detection of MDR strains in these hospitals and enable the prescription of suitable treatment for the patient [8]. Since it demonstrates higher sensitivities and specificities than other methods, REMA can help reduce the incidence of TB and DR-TB. In addition, we plan to use REMA for the DST of other first-line and second-line anti-TB drugs to detect extensively drug-resistant TB at an early stage. The results of the present study may contribute to reducing the rate of TB and drug-resistant TB by demonstrating the high sensitivities and specificities of REMA. Our results are compared to the results of other authors in Table 3 [8,9,13,20−23].
- Genotypic DST by RT-PCR TaqMan Allelic Discrimination Assay
- Although conventional DST is considered the “gold standard” for assessing DR-TB, its disadvantages include slow turnaround times and high rates of contamination. In contrast, molecular assays offer faster results without compromising sensitivity and specificity [24,25]. In this study, we used the RT-PCR TaqMan allelic discrimination assay to test 52 M. tuberculosis clinical isolates, including 31 MDR, 1 RIF mono-resistant, 10 INH mono-resistant, and 10 drug-sensitive isolates. Previous research has shown that drug-resistant strains of M. tuberculosis can be detected by observing curve patterns or Ct values using 3 TaqMan probes (without minor groove binder) in an RT-PCR format [14]. The proposed method was optimized and evaluated for analytical sensitivity and specificity in the clinical isolates. When compared with DNA sequencing, this method provided good sensitivity and specificity in the results of our study and of other studies [26,27]. Since there is a lack of information on the genetic characteristics of M. tuberculosis isolates in central Vietnam, the use of molecular DST could aid in identifying the diversity of drug resistance-associated mutation patterns and provide insights into the genotypic DR-TB isolates in Vietnam. This will support a focus on infection control and surveillance to prevent new cases of MDR-TB in the region.
- In this study, sequencing of the katG gene confirmed the presence of the Ser315Thr (AGC>ACC) mutation in 19 of 41 (46.3%) INH-resistant isolates, including 15 of 31 (48.4%) MDR-TB isolates and 4 of 10 (40.0%) INH mono-resistant isolates. This study only investigated mutations from codon 256 to codon 420 of the katG gene, and Ser315 was found to be the most frequently encountered mutation associated with INH resistance. This is consistent with other research findings such as a systematic review of 118 publications analyzing 11,411 M. tuberculosis isolates from 49 countries, which found that 64% of all observed phenotypic INH resistance was associated with mutation katG315 [28]. A study by Chikamatsu et al. [29] reported that 54.5% of isolates harbored the katG315 mutation in Japan. In addition, van Doorn et al. [30] found that 55% of INH-resistant strains had a mutation in codon 315 of katG, while Riahi et al. [27], reported approximately 53.55%.
- In a recent study by Sadri et al. [31] in Iran, of 125 M. tuberculosis clinical isolates, 34 strains were INH-resistant, and 91 strains were INH-sensitive. Of the 34 INH-resistant strains, 32% of the mutations were identified in katG315 (Ser→Thr), 14% in katG315 (Ser→ Asn), 52% in InhA 15 (C→T), and 2.9% in InhA 17 (G→T).
- The question remains, which genetic variants in INH-resistant bacteria are related to the prevalence of genetic mutations conferring M. tuberculosis drug resistance in these different geographic regions. INH resistance was found to be more difficult to detect because the mutations were associated with multiple genes. In our study, we only investigated katG at codon 315. Further studies are needed to gain a complete understanding of the genetic variations in M. tuberculosis resistance to INH drugs, and to determine the prevalence of resistant mutations among M. tuberculosis isolates in the different geographic regions of Vietnam.
- We utilized 2 wild-type probes for the rpoB gene to determine RIF resistance in this region, including the point mutations mentioned above. The most frequently detected mutation was rpoB Ser531Leu (TCG>TTG) at 37.5%. We identified 4 different types of amino acid substitution in codon 526, with His526Asp (CAC>GAC) mutations present in 2 isolates, and His526Tyr (CAC>TAC) mutations present in 2 isolates. One isolate had a His256Asn (CAC>AAC) mutation, and 2 isolates had His256Pro (CAC>CCC) mutations. Only 2 strains (6.25%) had mutations at codon Asp516Tyr (GAC>TAC). One strain had a double point mutation in L511Pro (CTG>CCG) and Met515Val (ATG>GTG). Only 1 case of RIF mono-resistance (0.21%) was detected, with a mutation in rpoB S531L. In a recent study in southern Vietnam, 56 isolates (approximately 76%) had mutations in the rifampin resistance-determining region (RRDR) of the rpoB gene, at codons 531 (37.8%), 526 (23%), and 516 (9.46%). In other research, 104 RIF-resistant isolates had mutations in the 81bp RRDR of the rpoB gene; the most prevalent mutations were at codons 531 (43%), 526 (31%), and 516 (15%) [32−34]. In the west of Iran, a study by Hamed et al. [35] found 32 MDR isolates among 54 M. tuberculosis samples, while the other 22 isolates were INH and RIF sensitive. Among the 32 MDR isolates, 22 strains (70%) had katG codon 315 mutations, 1 strain (3%) had inhA mutations, and 9 strains (28%) had no mutations at all. Regarding RIF, mutations associated with the rpoB gene in codons 531, 516 and 626 comprised 21 strains (66%), 7 strains (22%) and 1 strain (3%) respectively, and no mutations were found in 3 strains (9%).
- In another study by Mohajeri et al. [36], 35 (28.0%) of 125 M. tuberculosis isolates were found to be RIF-resistant with S512T (AGC>ACC; 20%), D516V (GAC>GTC; 20%), H526D (CAC>GAC; 6%), H526R (CAC>CGC; 20%), H526Y (CAC>TAC; 23%), and S531W (TCG>TGG; 8%), and the most frequent site mutations were L511P (CCG>CTG; 46%), followed by S531l (TCG>TTG; 40%) and D516Y (GAC>TAC; 26%). This study detected resistance in strains with mutations within the hotspot at 81 bp but did not find cases with mutations located outside the hot spot of rpoB. Our study could only examine a short segment of the rpoB and did not observe the rpoA and rpoC of M. tuberculosis.
Discussion
- This study highlights the importance of developing molecular tests that allow quick screening for INH and RIF resistance-related mutations in Vietnam, where a significant percentage of patients with TB remain undiagnosed and untreated. The use of the RT-PCR TaqMan allelic assays, followed by the determination of MIC values using REMA, can assist clinicians when deciding on effective drug resistance regimens for their patients with DR-TB. This approach can help control the spread of MDR-TB, not only in Vietnam but globally as well. Therefore, it is recommended that laboratories in pulmonary hospitals adopt this approach to improve the diagnosis and treatment of DR-TB patients.
Conclusion
- Tuberculosis (TB) and drug-resistant TB (DR-TB) are serious public health problems. New strategies are urgently needed to manage and reduce the mortality related to this disease, especially in resource-limited settings. To prevent new cases of DR-TB in Vietnam, a comprehensive understanding of the genotypes of DR-TB isolates is crucial so that the most appropriate and effective diagnostic methods for detecting DR-TB can be developed and implemented.
HIGHLIGHTS
-
Ethics Approval
Ethical approval for this study was granted by The Ethical Committee in Biomedical Research of the University of Medicine and Pharmacy-Hue University (No: H2019/350). The director of each hospital granted permission to conduct the study. Before specimen collection, patients/guardians were provided with information about the study and asked to provide written informed consent. Participants were informed of their right to voluntarily participate in the study.
-
Conflicts of Interest
The authors have no conflicts of interest to declare.
-
Funding
This study was also supported by project No. DHH 2020-04-111 of Hue University, Hue, Vietnam.
-
Availability of Data
All data generated or analyzed during this study are included in this published article. Other data may be requested through the corresponding author.
-
Authors’ Contributions
Conceptualization: TBNN, VQTN, PM; Data curation: HBN, vBTP, TKDN; Formal analysis: VQTN, VTM, TBNN; Funding acquisition: all authors; Methodology: PM, VQTN, TBNN; Project administration: TBNN, PM, TTN; Visualization: PM, VQTN, VTM, TBNN; Writing–original draft:TBNN, VQTN; Writing–review & editing: all authors. All authors read and approved the final manuscript.
-
Additional Contributions
The authors would like to thank all faculty members, residents, and technical staff members at the Department of Microbiology and Clinical Microbiology, Hospital University Sassari, Italy.
Article information
-
Acknowledgements
- This work was also supported by professors at Sassari University, Department of Microbiology and Clinical Microbiology, Hospital University Sassari, Italy.

| History | Overall (%) | INH (%) | RIF | MDR-TB (%) |
|---|---|---|---|---|
| New TB cases | 413 (88.2) | 89 (21.6) | 0 | 23 (5.6) |
| Previous TB cases | 55 (11.8) | 16 (29.1) | 1 | 8 (14.5) |
| p | 0.006 | 0.208 | NA | 0.012 |
| Study | Country | No. of clinical isolates | Reference test | Sample size (no. of resistant/no. of susceptible) | Sensitivity (95% CI) | Specificity (95% CI) |
|---|---|---|---|---|---|---|
| INH | ||||||
| Palomino et al. [9] (2002) | Belgium | 80 | LJ | 54/26 | 1.00 (0.93–1.00) | 0.96 (0.80–1.00) |
| Martin et al. [13] (2005) | Belgium | 203 | LJ | 82/212 | 0.98 (0.91–1.00) | 0.98 (0.93–0.99) |
| Nateche et al. [8] (2006) | Algeria | 136 | LJ | 17/119 | 1.00 (0.80–1.00) | 0.99 (0.95–1.00) |
| Coban et al. [21] (2006) | Turkey | 73 | BACTEC | 35/38 | 1.00 (0.85–1.00) | 0.95 (0.76–0.99) |
| Dixit et al. [22] (2012) | India | 105 | LJ | 51/54 | 0.93 | 0.98 |
| Nour et al. [23] (2013) | Egypt | 30 | PM | 20/10 | 1.00 | 0.98 |
| This study | Vietnam | 468 | BACTEC | 105/363 | 100 (0.96–1.00) | 0.99 (0.98–1.00) |
| RIF | ||||||
| Palomino et al. [9] (2002) | Belgium | 80 | LJ | 49/31 | 1.00 (0.93–1.00) | 1.00 (0.89–1.00) |
| Martin et al. [13] (2005) | Belgium | 203 | LJ | 102/101 | 0.98 (0.93–1.00) | 0.99 (0.95–1.00) |
| Nateche et al. [8] (2006) | Algeria | 136 | LJ | 12/124 | 0.92 (0.62–1.00) | 0.99 (0.96–1.00) |
| Coban et al. [21] (2006) | Turkey | 73 | BACTEC | 21/52 | 1.00 (0.81–1.00) | 0.94 (0.89–1.00) |
| Dixit et al. [22] (2012) | India | 105 | LJ | 52/53 | 0.95 | 1.00 |
| Nour et al. [23] (2013) | Egypt | 30 | PM | 13/17 | 0.95 | 0.93 |
| This study | Vietnam | 468 | BACTEC | 32/436 | 0.99 (0.88–1.00) | 0.94 (0.98–0.99) |
- 1. Davies PD, Lalvani A, Thillai M, et al. Clinical tuberculosis: a practical handbook. CRC Press; 2016.
- 2. Tattersfield A. Toman’s tuberculosis: case detection, treatment and monitoring: questions and answers, 2nd edition. Occup Environ Med 2005;62:70.
- 3. Daniel TM. The history of tuberculosis. Respir Med 2006;100:1862−70.ArticlePubMed
- 4. World Health Organization (WHO). Global tuberculosis report. WHO; 2020.
- 5. Rich M; World Health Organization (WHO). Guidelines for the programmatic management of drug-resistant tuberculosis Guidelines for the programmatic management of drug-resistant tuberculosis. WHO; 2006.
- 6. Nhung NV, Hoa NB, Sy DN, et al. The fourth national anti-tuberculosis drug resistance survey in Viet Nam. Int J Tuberc Lung Dis 2015;19:670−5.ArticlePubMed
- 7. Heifets L. Mycobacteriology laboratory. Clin Chest Med 1997;18:35−53.ArticlePubMed
- 8. Nateche F, Martin A, Baraka S, et al. Application of the resazurin microtitre assay for detection of multidrug resistance in Mycobacterium tuberculosis in Algiers. J Med Microbiol 2006;55(Pt 7). 857−60.ArticlePubMed
- 9. Palomino JC, Martin A, Camacho M, et al. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2002;46:2720−2.ArticlePubMedPMCPDF
- 10. Cirillo DM, Miotto P, Tortoli E. Evolution of phenotypic and molecular drug susceptibility testing. Adv Exp Med Biol 2017;1019:221−46.
- 11. Menzies D, Benedetti A, Paydar A, et al. Standardized treatment of active tuberculosis in patients with previous treatment and/or with mono-resistance to isoniazid: a systematic review and meta-analysis. PLoS Med 2009;6:e1000150.ArticlePubMedPMC
- 12. Almeida Da Silva PE, Palomino JC. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. J Antimicrob Chemother 2011;66:1417−30.ArticlePubMed
- 13. Martin A, Morcillo N, Lemus D, et al. Multicenter study of MTT and resazurin assays for testing susceptibility to first-line anti-tuberculosis drugs. Int J Tuberc Lung Dis 2005;9:901−6.PubMed
- 14. Darban-Sarokhalil D, Nasiri MJ, Fooladi AA, et al. Rapid detection of rifampicin- and isoniazid-resistant Mycobacterium tuberculosis using TaqMan allelic discrimination. Osong Public Health Res Perspect 2016;7:127−30.ArticlePubMedPMC
- 15. Wada T, Maeda S, Tamaru A, et al. Dual-probe assay for rapid detection of drug-resistant Mycobacterium tuberculosis by real-time PCR. J Clin Microbiol 2004;42:5277−85.ArticlePubMedPMCPDF
- 16. Martin A, Camacho M, Portaels F, et al. Resazurin microtiter assay plate testing of Mycobacterium tuberculosis susceptibilities to second-line drugs: rapid, simple, and inexpensive method. Antimicrob Agents Chemother 2003;47:3616−9.ArticlePubMedPMCPDF
- 17. Stagg HR, Lipman MC, McHugh TD, et al. Isoniazid-resistant tuberculosis: a cause for concern? Int J Tuberc Lung Dis 2017;21:129−39.ArticlePubMed
- 18. Jagielski T, Bakuła Z, Roeske K, et al. Detection of mutations associated with isoniazid resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates. J Antimicrob Chemother 2014;69:2369−75.ArticlePubMed
- 19. Gagneux S, Long CD, Small PM, et al. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 2006;312:1944−6.ArticlePubMed
- 20. Miyata M, Pavan FR, Sato DN, et al. Comparison of resazurin microtiter assay performance and BACTEC MGIT 960 in the susceptibility testing of Brazilian clinical isolates of Mycobacterium tuberculosis to four first-line drugs. Braz J Microbiol 2013;44:281−5.ArticlePubMedPMC
- 21. Coban AY, Cekic Cihan C, Bilgin K, et al. Rapid susceptibility test for Mycobacterium tuberculosis to isoniazid and rifampin with resazurin method in screw-cap tubes. J Chemother 2006;18:140−3.ArticlePubMed
- 22. Dixit P, Singh U, Sharma P, et al. Evaluation of nitrate reduction assay, resazurin microtiter assay and microscopic observation drug susceptibility assay for first line antitubercular drug susceptibility testing of clinical isolates of M. tuberculosis. J Microbiol Methods 2012;88:122−6.ArticlePubMed
- 23. Nour MS, El-Shokry MH, Shehata IH, et al. Evaluation of rezasurin microtiter assay and high resolution melting curve analysis for detection of rifampicin and isoniazid resistance of Mycobacterium tuberculosis clinical isolates. Clin Lab 2013;59:763−71.ArticlePubMed
- 24. Helb D, Jones M, Story E, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol 2010;48:229−37.ArticlePubMedPDF
- 25. Nonghanphithak D, Kaewprasert O, Chaiyachat P, et al. Whole-genome sequence analysis and comparisons between drug-resistance mutations and minimum inhibitory concentrations of Mycobacterium tuberculosis isolates causing M/XDR-TB. PLoS One 2020;15:e0244829.ArticlePubMedPMC
- 26. Peng J, Yu X, Cui Z, et al. Multi-fluorescence real-time PCR assay for detection of RIF and INH resistance of M. tuberculosis. Front Microbiol 2016;7:618. ArticlePubMedPMC
- 27. Riahi F, Derakhshan M, Mosavat A, et al. Evaluation of point mutation detection in Mycobacterium tuberculosis with isoniazid resistance using real-time PCR and TaqMan probe assay. Appl Biochem Biotechnol 2015;175:2447−55.ArticlePubMedPDF
- 28. Seifert M, Catanzaro D, Catanzaro A, et al. Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: a systematic review. PLoS One 2015;10:e0119628.ArticlePubMedPMC
- 29. Chikamatsu K, Mizuno K, Aono A, et al. Evaluation of GenoType MTBDRplus for the detection of multi-drug-resistant Mycobacterium tuberculosis strains. Kekkaku 2011;86:697−702. Japanese.PubMed
- 30. van Doorn HR, Claas EC, Templeton KE, et al. Detection of a point mutation associated with high-level isoniazid resistance in Mycobacterium tuberculosis by using real-time PCR technology with 3'-minor groove binder-DNA probes. J Clin Microbiol 2003;41:4630−5.ArticlePubMedPMCPDF
- 31. Sadri H, Farahani A, Mohajeri P. Frequency of mutations associated with isoniazid-resistant in clinical Mycobacterium tuberculosis strains by low-cost and density (LCD) DNA microarrays. Ann Trop Med Public Health 2016;9:307−11.Article
- 32. Minh NN, Van Bac N, Son NT, et al. Molecular characteristics of rifampin- and isoniazid-resistant Mycobacterium tuberculosis strains isolated in Vietnam. J Clin Microbiol 2012;50:598−601.ArticlePubMedPMCPDF
- 33. Tho DQ, Ha DT, Duy PM, et al. Comparison of MAS-PCR and GenoType MTBDR assay for the detection of rifampicin-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis 2008;12:1306−12.PubMed
- 34. Caws M, Duy PM, Tho DQ, et al. Mutations prevalent among rifampin- and isoniazid-resistant Mycobacterium tuberculosis isolates from a hospital in Vietnam. J Clin Microbiol 2006;44:2333−7.ArticlePubMedPMCPDF
- 35. Hamed Z, Mohajeri P, Farahani A, et al. The frequency of point mutations associated with resistance to isoniazid and rifampin among clinical isolates of multidrug-resistant Mycobacterium tuberculosis in the west of Iran. Gene Rep 2021;22:100981. Article
- 36. Mohajeri P, Sadri H, Farahani A, et al. Frequency of mutations associated with rifampicin resistance in Mycobacterium tuberculosis strains isolated from patients in west of Iran. Microb Drug Resist 2015;21:315−9.ArticlePubMed
References
Figure & Data
References
Citations



 Cite
Cite