Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 13(6); 2022 > Article
-
Review Article
SARS-CoV-2 in brief: from virus to prevention -
Hassan Karami1
 , Zeinab Karimi1
, Zeinab Karimi1 , Negin Karami2
, Negin Karami2
-
Osong Public Health and Research Perspectives 2022;13(6):394-406.
DOI: https://doi.org/10.24171/j.phrp.2022.0155
Published online: November 28, 2022
1Department of Virology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran
2Department of Nursing, School of Nursing, Alborz University of Medical Sciences, Karaj, Iran
- Corresponding author: Hassan Karami Department of Virology, School of Public Health, Tehran University of Medical Sciences, Poursina Avenue, Qods Street, Enqelab Square, Tehran, Iran E-mail: Karami.hassan.2022@gmail.com
© 2022 Korea Disease Control and Prevention Agency.
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Abstract
- The recent outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a highly transmissible virus with a likely animal origin, has posed major and unprecedented challenges to millions of lives across the affected nations of the world. This outbreak first occurred in China, and despite massive regional and global attempts shortly thereafter, it spread to other countries and caused millions of deaths worldwide. This review presents key information about the characteristics of SARS-CoV-2 and its associated disease (namely, coronavirus disease 2019) and briefly discusses the origin of the virus. Herein, we also briefly summarize the strategies used against viral spread and transmission.
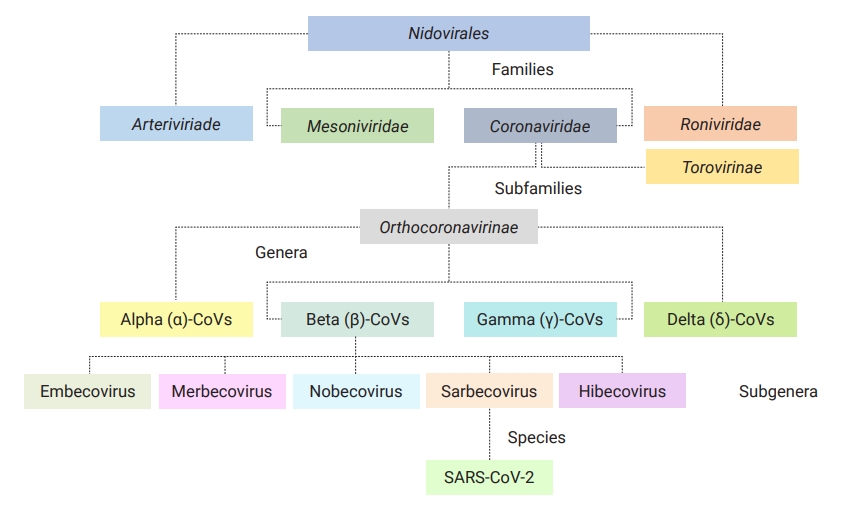
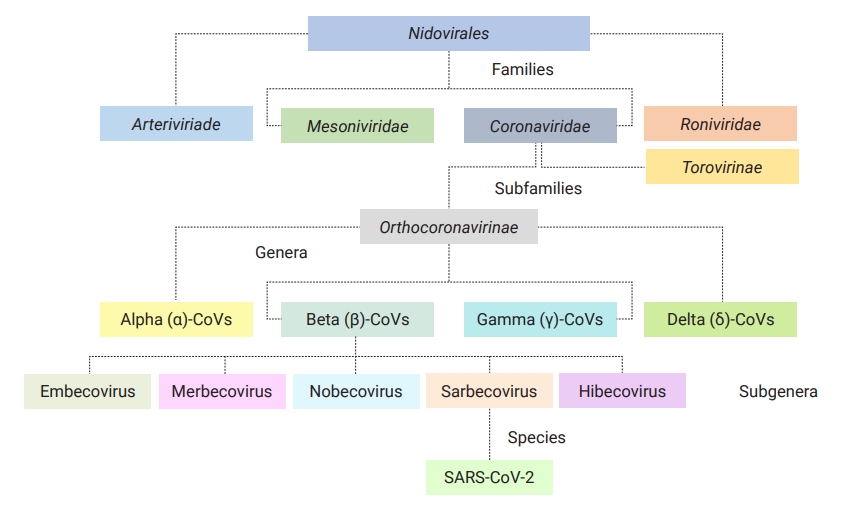
- Coronaviruses (CoVs) are classic, small infectious agents in both humans and animals [1]. Belonging to the family of Coronaviridae in the order Nidovirales, these viruses are divided into 2 subfamilies (Orthocoronavirinae and Torovirinae) [1]. Having 4 distinct genera in 4 major subclades, the subfamily of Orthocoronavirinae is classified into alpha (α)-CoVs and beta (β)-CoVs (both are infectious for mammals), as well as gamma (γ)-CoVs (infectious for birds), and delta (δ)-CoVs (infectious for mammals and birds) sharing remarkable interspecies similarities; however, differences in some features such as genome composition, transmission, pathogenicity, and associated diseases are notable (Figure 1) [1,2]. Before December 2019, 6 pathogenic CoVs had been identified as causing mild to severe human respiratory infections. Of these, 4 viruses were low-pathogenic (HCoV-OC43, HCoV-NL63, HCoV-HKU1, and HCoV-229E) associated with mild infections, and 2 viruses (severe acute respiratory syndrome coronavirus 1 [SARS-CoV-1; 2002–2003, Foshan in China] and Middle East respiratory syndrome coronavirus [MERS-CoV; 2012, Arabian Peninsula]) were highly pathogenic and linked to severe acute respiratory syndrome [3–5].
- On December 31, 2019, the World Health Organization (WHO) received an alert from China and was informed of a cluster of unexplained pneumonia [6]. A week later, a new strain of CoV was detected and subsequently announced by officials of the same organization [6]. Soon after, the WHO and the International Committee on Taxonomy of Viruses chose, respectively, the names 2019 novel coronavirus (2019-nCoV) and SARS-CoV-2 for the newly isolated virus [7]. Historically, SARS-CoV-2 was found after an outbreak of an unknown airway infectious disease in the city of Wuhan, China [6]. Shortly afterward, the outbreak made headlines in both regional and global news in early 2020 and plunged the world into a state of concern, and was, in fact, the beginning of a health crisis on a worldwide scale. Due to its rapid spread, the outbreak was declared a public health emergency of international concern and was deemed to constitute a global pandemic on January 30, 2020 and March 11, 2020, respectively [8].
- SARS-CoV-2 is the latest infectious CoV to be identified in humans; it causes mildly symptomatic disease in about 80% of infected cases, but adverse outcomes are also predictable in the 15% and 5% of patients who develop severe and critical disease, respectively [9]. As of September 21, 2022, there have been more than 618 million confirmed coronavirus disease 2019 (COVID-19) cases, more than 6.5 million deaths, and nearly 600 million recoveries worldwide (https://www.worldometers.info/coronavirus/). To control the transmission of COVID-19 and curtail the associated infections, we should enhance our knowledge regarding the characteristics and origin of SARS-CoV-2, as well as the mechanisms underlying the associated disease. There is also a need to focus on interventions to reduce the rate of viral transmission, infection, and death.
Introduction
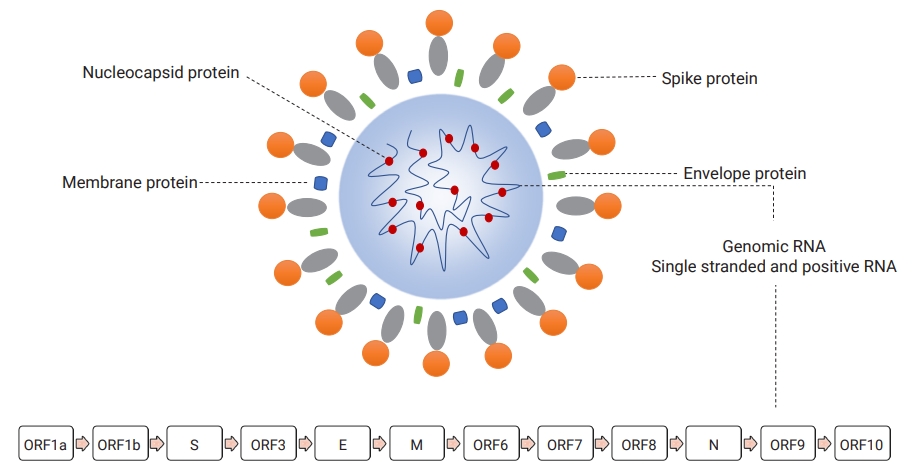
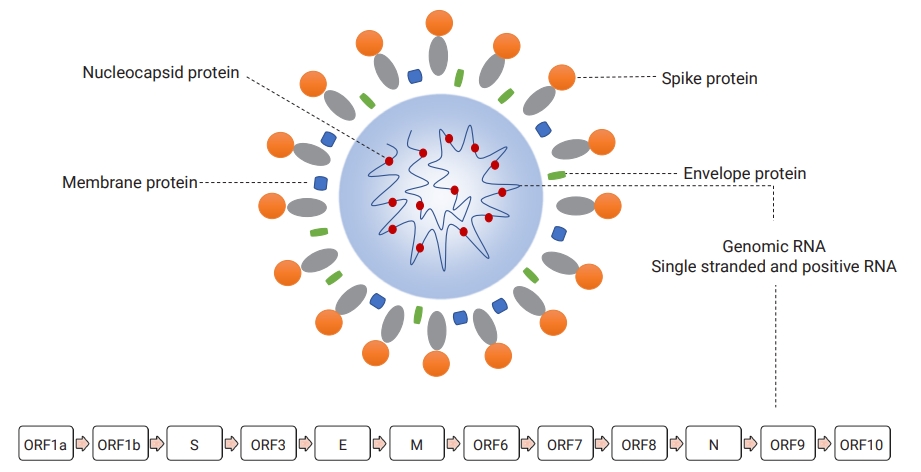
- Morphologically, early microscopic observations found the SARS-CoV-2 virion as a pleomorphic, rounded, and crown-shaped particle measuring 60 to 140 nm in diameter [10]. This virus contains a helical nucleocapsid protein that directly interacts with an intraparticle genomic nucleic acid as a complex of ribonucleoprotein, which are surrounded by a double-layered lipid originally derived from the membrane of infected cells [11]. In addition, the outer membrane of infectious viruses contains several projected proteins (referred to as spike proteins), which are 9 to 12 nm in diameter (Figure 2) [10].
- Like other CoVs, SARS-CoV-2 has a capped and polyadenylated, large, constant, single, and positive-sense ribonucleic acid organized in a specific order into different gene sequences responsible for 3 general types of functional proteins [12]. The SARS-CoV-2 RNA encodes at least 29 different functional proteins essential for viral replication, infectivity, immunomodulation, and future therapeutic and vaccine research [13]. The genome of SARS-CoV-2 contains 2 overlapping regions (open reading frame-1a [ORF1a] and ORF1b) encoding 16 non-structural proteins (NSP1-16). In addition, it encodes 4 structural proteins (spike [S], envelope [E], membrane [M], and helical nucleocapsid [N]) and a set of accessory proteins (ORF3, ORF6, ORF7, ORF8, ORF9, ORF10) with a variable length, ranging from 13 amino acids to 1945 amino acids for NSP11 and NSP3, respectively (Figure 2) [13,14].
- The nucleic acid sequence of SARS-CoV-2, with a high sequence identity, is closely related to bat coronavirus RaTG13 (96.10%), RpYN06 (94.48%), RmYN02 (93.30%), SARS-like‐CoVZC45 (87.60%), SARS-like‐CoVZXC21 (87.40%), SARS-CoV-1 (79.6%), and MERS-CoV (50%) [15–17]. Despite these similarities, SARS-CoV-2 differs from SARS-CoV-1 genetically. For instance, in SARS-CoV-2, ORF8b is longer (37 amino acids) and ORF3b is shorter (132 amino acids) than in SARS-CoV-1. In addition, there are no genes encoding either ORF8a or hemagglutinin esterase in SARS-CoV-2 [18,19].
- The genome of SARS-CoV-2 undergoes variation, deletion, insertion, and broad mutations (estimated approximately 1×10−3 substitutions per year) leading to the emergence of new lineages/variants with new viral and infectious characteristics [20]. To date, 3 classifications—variant of concern (VOC), variant of interest (VOI), and variant under monitoring (VUM)—have been introduced for variants of SARS-CoV-2. The first term (VOC) refers to variants with high transmissibility that can be highly virulent, with a negative impact on vaccine efficiencies and therapeutic prospects [21]. Currently, these variants are challenging to human health. The second term (VOI) denotes variants that harbor genetic mutations predicted to have an impact on viral transmissibility and disease severity [22]. The last term (VUM) refers to variants that are genetically changed and are likely to pose a threat in the future due to their unknown phenotypic or epidemiological effects [21]. Lists of variants, countries, and selected virus spike mutations are summarized in Table 1.
The Virus: Structure, Genetics, Variants
- The recent literature still contains no clear data to answer the question of whether SARS-CoV-2 is a natural or man-made virus; however, research on this mysterious topic is still being undertaken. Almost all human CoVs are thought to be of animal origin, as these viruses can spread to humans by cross-species transmission [23]. The SARS-CoV-2 pandemic seems to have started in the Wuhan wet market in China, where different kinds of animals were traded [24]. Bats and pangolins are likely the natural hosts of SARS-CoV-2 [25,26]. However, for more than 2 years before the start of the current pandemic, those animals were not for sale in the Wuhan wet market [24]. Therefore, additional hosts (reservoirs and intermediate ones) may need to be identified through further investigations.
- To date, different scenarios have been described regarding the origin of SARS-CoV-2. In this context, 2 scenarios (zoonotic and laboratory origin) are thought to be more likely than the others. Briefly, the first one is supported by genomic data showing the same mutations in 6 residues of viral receptor-binding domains (RBDs) in both SARS-CoV-2 and pangolin-derived CoVs, suggesting the appearance of natural selection in pangolins before its transfer to humans [27]. In addition, the spike protein of SARS-CoV-2 includes a furin polybasic cleavage site and o-linked glycans, which both seem to have been naturally generated, likely through immunity-induced pressure [27].
- In contrast to the natural selection scenario, it has been assumed that SARS-CoV-2 was artificially synthesized under controlled conditions by a combination of RaTG13‐like and MP789‐ like CoVs [28]. This assumption was discussed in a recently published letter. In this letter, no sufficient data were provided in favor of the man-made origin of SARS-CoV-2 [29]. In another claim, SARS-CoV-2 was stated to be a recombinant virus having an inserted fragment (1,387 bp) corresponding to the viral spike protein; however, this statement was soon refuted by a group of researchers who found that this sequence was not unique to SARS-CoV-2, since other CoVs clearly showed the same genetic pattern in their spike proteins [30]. However, on March 30, 2021, a group of experts reported that although laboratory leakage could possibly have occurred, for SARS-CoV-2, it is an “extremely unlikely” route [31]. In general, more studies should be done to find a definitive answer to the question of whether SARS-CoV-2 is a naturally selected or laboratory-generated/leaked virus.
Origin
- It is well-established that being unprotected and close to a person infected with SARS-CoV-2 regardless of whether he or she is symptomatic or not, increases the risk of viral transmission, especially in communities with high levels of interpersonal contact [6,32]. This mode of transmission (interpersonal transmission) which was first confirmed on January 20, 2020, highlights the importance of taking appropriate precautions when attending public gatherings outside the home [8]. Horizontally, expiratory activities generate up to a few million droplets of oral fluids with sizes of <1 to 1,000 µm [33,34]. Large droplets (60–100 µm) are formed and expelled into the air primarily by defensive reflexes of the respiratory system, which throw virus-laden liquid particles up to a distance of approximately 1 meter for exhalation to 2 meters for coughing and 6 meters for sneezing within 0.12 to 1 second in a velocity of 1 to 50 m/s [33].
- SARS-CoV-2 is a contagious virus and, indeed, what has made this virus a more transmissible CoV strain than SARS-CoV-1 and MERS-CoV might be the number of detectable viruses which are shed outside the mouse/nose [34]. SARS-CoV-2-laden aerosols suspended in the air were found to travel for distances up to 4 meters and are similarly infectious to SARS-CoV-1 at a median interval of an hour after generation, increasing the likelihood of infection if inhaled or deposited on the mucosa [32,35,36]. Being in an enclosed environment (i.e., restaurants, classrooms, gyms, or prisons) where there is no proper or poor air circulation may facilitate the virus spread, increasing the risk of interhuman viral infection [37]. Although the minimum number of virions required to start an infection has not been reported, early studies estimated that around 100 infectious particles are enough to infect those who are not appropriately protected against viral transmission [38].
- In indirect transmission, SARS-CoV-2 may spread through inanimate surfaces. This may occur through touching cross-contaminated or directly contaminated commonly used objects [39,40]. However, this mode of transmission is considered less important than direct contact for viral spread and transmission [41]. Nonetheless, various environmental conditions such as the level of light, pH, ultraviolet irradiation, temperature, and humidity need to be set to find how much these parameters affect the stability of the virus on contaminated surfaces. In addition to all the above routes, infection may also occur by exposure to virus-containing non-respiratory biofluids since molecular detection has confirmed the presence of viral nucleic acid in human body fluids such as breast milk, amniotic fluid, blood products, sexual secretions, and urinary and gastrointestinal excretions in SARS-CoV-2-infected individuals [42–45]. This raises concerns regarding additional modes of virus transmission.
Transmission
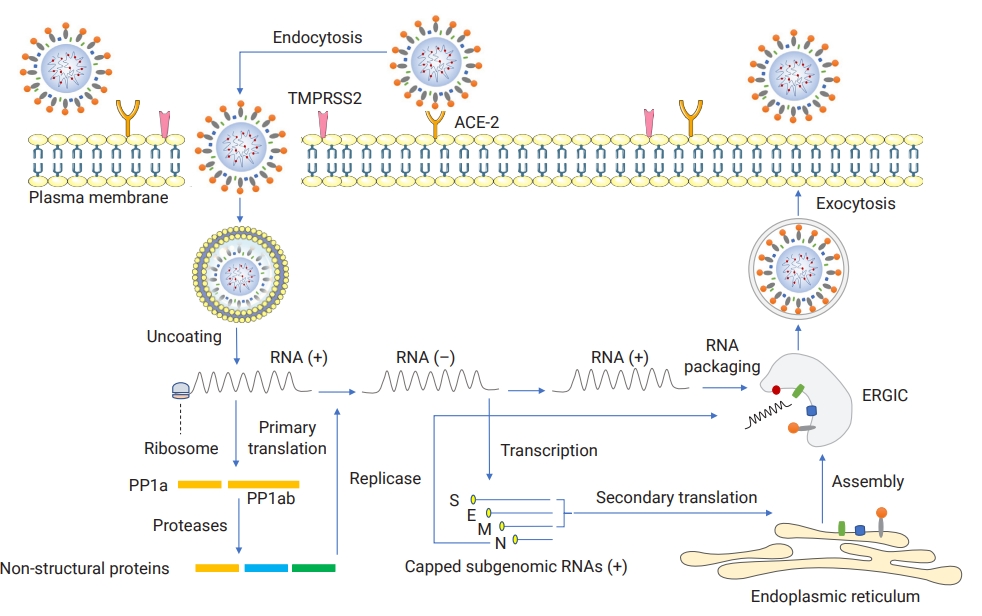
- Basic information exists on the pathogenic mechanisms of SARS-CoV-2 and the mechanisms underlying the progression of the disease to unfavorable outcomes. SARS-CoV-2 encodes a trimeric, mutable, and surface class I viral fusion protein called the S glycoprotein, which consists of S1 (binding) and S2 (anchoring) subunits associated together non-covalently [46]. The S1 subunit harbors a functional and antigenic domain called the RBD, which is responsible for receptor engagement in virus-cell interaction, whereas S2 fuses viral and cell membranes to help the virus enter the cell [46]. The spike protein of SARS-CoV-2 is remarkably similar (approximately 78% identity in amino acid sequences) to its equivalent presented on the SARS-CoV-1 membrane [47]. SARS-CoV-2 is a novel airway-associated infectious agent that primarily affects the host respiratory system and likely exhibits systemic involvement with non-respiratory systems due to its broad tissue tropism explained by the wide expression pattern of angiotensin-converting enzyme-2 (ACE-2) as the predominant cellular receptor (also for SARS-CoV-1) and its co-expressed molecule, the transmembrane serine protease 2, as a cellular and spike priming protease [48,49]. These molecules mediate the virus attachement penetration through the endocytic pathway, which is followed by key replicative steps that eventually form viral particles and lead to the release of new infectious particles out of the infected cell (Figure 3) [14]. For SARS-CoV-2, additional and alternative molecules have recently been proposed to serve as entry mediators [46,50,51]. In contrast to ACE-2, dipeptidyl-peptidase 4 and aminopeptidase N are often utilized by MERS-CoV and HCoV-229E, respectively, to invade the cells of choice [46].
- As mentioned above, SARS-CoV-2 is a respiratory virus, meaning that the respiratory system might be the primarily affected organ system during infection [52]. Following the inhalation of virus-laden particles, the infection begins by attacking the epithelium of the respiratory tract primarily via targeting nasal multiciliated and sustentacular cells as well as oral glands, and mucous membranes enriched by cell-associated SARS-CoV-2 entry molecules [53–56]. Inside invaded cells, the viruses are sensed via cytoplasmic recognition molecules, which result in interferon (IFN)-mediated innate immune responses derived from the activation of IFN regulatory factors 3 and 7 (IRF3 and IRF7) [51].
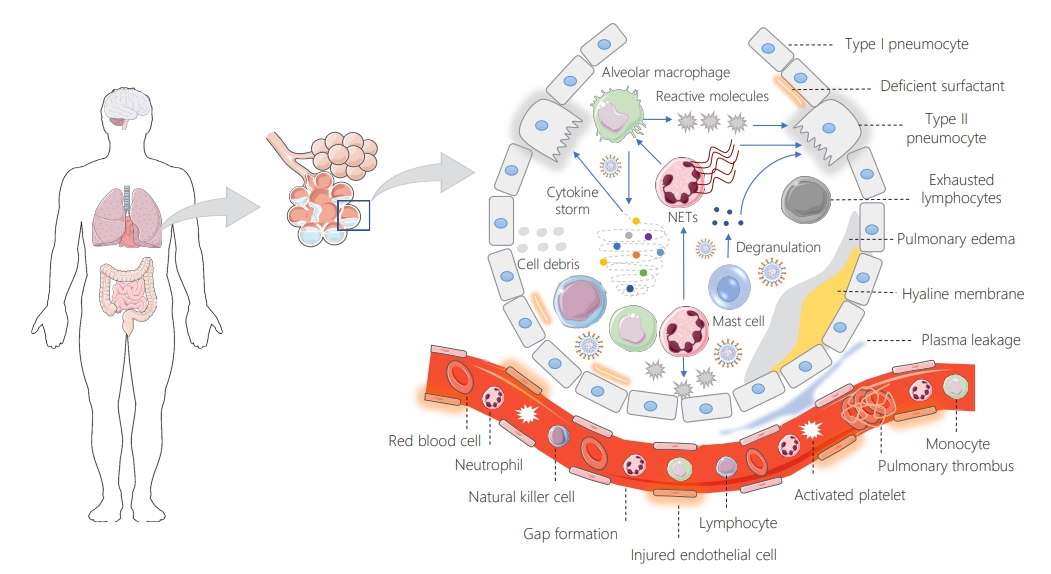
- As shown in Figure 4, by reaching the lower airway (alveoli) through the conducting airways, SARS-CoV-2 preferentially invades alveolar epithelial type II cells, allowing the virus to efficiently replicate, making more viruses and infect more cells [6]. Upon cell entry and virus replication, the activation of pattern recognition receptors and inflammatory signaling pathways initially results in cytokine and chemokine production [57]. However, SARS-CoV-2 has evolved to interfere with these intracellular recognition pathways [58]. When they fill the alveolar lumen, these inflammatory mediators mediate the recruitment of a subset of mono- and poly-morphonuclear blood cells into the site of infection, where the immune system responds to viruses that have entered [59]. The attracted leukocytes, predominantly monocyte-derived macrophages, contribute to the enhancement of host immune responses characterized by uncontrolled and storm-like cytokine activity [6,60]. The excessive release of cytokines results in capillary permeability and plasma leakage, and it also promotes further pulmonary inflammation and tissue injuries, which are associated with virus-induced acute respiratory distress syndrome, characterized by hypoxemia (impaired oxygenation) and organ deprivation of respiratory gases [9,61,62]. Furthermore, activated resident (e.g., macrophages) or recruited (e.g., neutrophils) immune cells contribute to injuries of both pneumocytes and endothelial barriers by inducing reactive molecules, which also play a role in defective T cell-mediated antiviral immunity [61,63]. SARS-CoV-2 induces neutrophil extracellular trap formation, which favors further inflammation triggered by macrophage-derived inflammatory cytokines [64]. The inflammation and alveolar injuries are also triggered by the rapid degranulation of mast cells, which are known as tissue-resident inflammation regulators cells [65]. SARS-CoV-2-infected cases whose lungs are seriously involved are at risk of alveolar collapse due to the loss of or low concentration of surfactant [66]. In these patients, hyaline membrane and intracapillary microthrombus formation are also expected [6]. Severe SARS-CoV-2 infection is also marked by impaired cell-mediated immunity, characterized by a numerical reduction (so-called lymphopenia) in the number of CD4+ and CD8+ T cells, Tregs, and γδ T cells and the functional exhaustion of peripheral PD-1+ and Tim-3+ expressing lymphocytes [64,67].
- As a pathophysiological mechanism, since the infection presents extrapulmonary manifestations, SARS-CoV-2 can infect cells outside the respiratory system, likely through hematogenous spread, leaving lesions in infected tissues and body organs [68].
Infection Mechanisms
- COVID-19 is a multifaceted and, more accurately, a multiphasic disease. While the entire course of the disease involves no or mild symptoms in most cases, severe to critical infections determined by organ involvement and the corresponding spectrum of clinical manifestations should also clinically be considered in a subset of SARS-CoV-2-infected individuals [8,69,70]. COVID-19 is a life-threatening disease, as it may lead to death at a median of 2 weeks after symptom onset; however, dying from the disease is relatively uncommon overall (case fatality rate, 1%) and most patients recover completely [8,56,71]. In this context, underlying comorbidities, such as non-communicable diseases, are among the risk factors for adverse outcomes such as hospitalization and death [72]. It was recently estimated that 84.1% of deaths due to COVID-19 occurred in patients with at least 1 medical condition [73]. Sex and age differences also affect the outcomes of infection in a non-favorable fashion [64].
- Through close and unprotected contact with confirmed patients, a symptomatic infection may develop within a week (median incubation period: 4 to 5 days) after virus exposure [56]. In early 2020, a brief report was published from a series of pneumonia patients who were clinically diagnosed with fever, cough, and chest discomfort [10]. Similar to the clinical pictures of other CoVs, the symptoms of COVID-19 may present in a combined pattern based on the disease severity [74]. Children with COVID-19 were found to be less likely to develop fever as a symptom of infection than adults [75]. A recent meta-analysis found medium-grade fever (ranging from 38.1°C to 39.0°C) in the majority of included patients, independent of the disease severity [75]. This symptom may last for 10 days on average in some hospital-admitted patients [76]. In these patients, the need for health care services may be associated with fever duration [77]. In the early stages of the disease, when the infection is mild, patients with acute SARS-CoV-2 infection may complain of muscle pain, headache, diarrhea, and most importantly, respiratory symptoms including nasal and throat congestion, coughing, rhinorrhea, sore throat, and shortness of breath at varying prevalence rates [78,79].
- As the disease progresses, the clinical symptoms show a moderate picture of severity approximately 1 week after symptom onset [56]. During this time, fever and coughing are likely to persist and breathlessness, tachypnea, moderate pneumonia, and abnormalities on chest computed tomography may manifest [78,80]. Coughing is a common sign that acutely presents in a non-productive pattern with no sputum production in the early days after illness onset, but as the disease progresses, the pattern changes to being productive [79]. In some patients, the infection is clinically characterized by severe and critical manifestations. In this stage, the infection towards organ dysfunction and failure, tissue injuries, severe pneumonia and dyspnea, hypoxia, cyanosis, and sepsis [78,80]. Symptomatic COVID-19 patients may also exhibit a variety of systemic symptoms, which are explained by the multiorgan involvement of the viral infection.
Clinical Manifestations
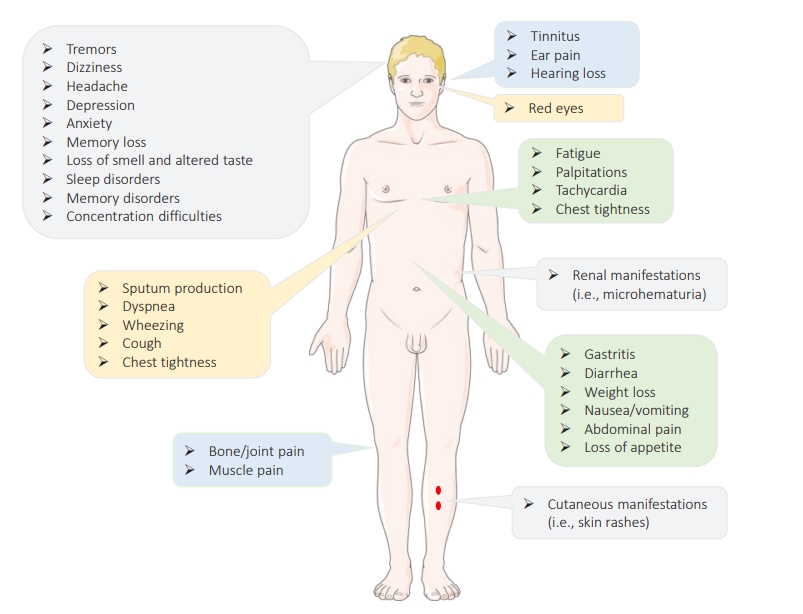
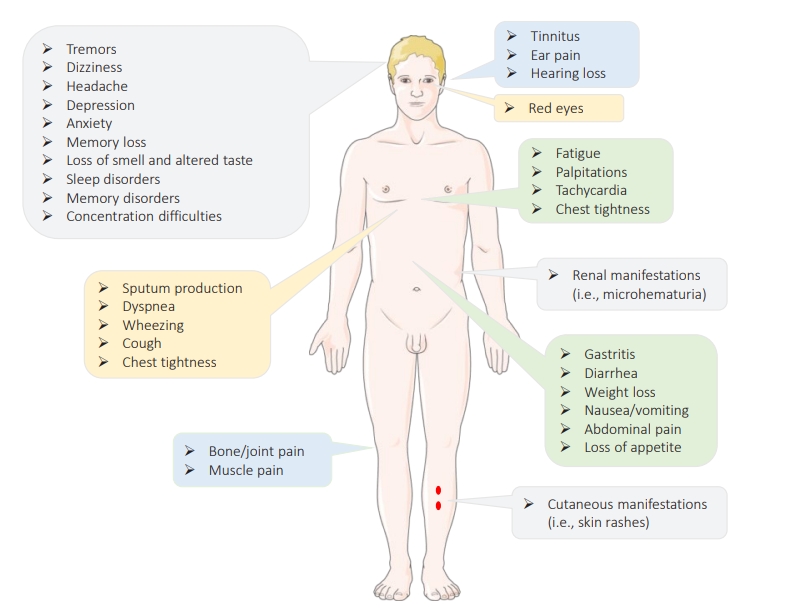
- The term “long COVID,” also known as “post-COVID-19 symptoms,” refers to a syndrome experienced by a subset (80%) of patients with a history of probable or confirmed COVID-19 who continue to experience for a long time (i.e., weeks after the acute infection) persistent symptoms (physical, mental, and/or cognitive symptoms) with no alternative diagnosis or explanation [81,82]. This syndrome is highly prevalent within the first 11 weeks after the onset of the disease, has a relapsing-remitting nature, and is not specific to SARS-CoV-2, since infection with other CoVs such as SARS-CoV-1 and MERS-CoV has been reported to have similar outcomes [82,83]. Long COVID appears to occur more likely in smokers, females, adults aged >35 years, those experiencing socioeconomic deprivation, patients with blood type A, people of White race, those who experienced hospital admission at the time of acute infection, patients with co-morbid conditions (e.g., asthma or obesity), and those with severe illness and poor general as well as mental health [83–88]. Patients with no or moderate symptoms at the time of acute infection are also at risk [89]. This syndrome is a multisystem disorder that can affect multiple body organs (e.g., the brain, skin, heart, kidney, lungs, etc.) characterized by different signs and symptoms (as summarized in Figure 5) [90,91]. While it is not clear how this syndrome is triggered, (1) viral-induced invasion and autoimmunity, (2) impaired immunometabolism, (3) immune exhaustion, (4) viral antigen persistence, (5) altered microbiome, (6) reactivation of latent viruses, and (7) increases and/or decreases in the renin-angiotensin system have been described as mechanisms that are likely involved in the pathophysiology of long COVID [92,93].
Long COVID
- Primary Prevention and Protection
- To mitigate the virus spread, prevent SARS-CoV-2 transmission, and end the current pandemic, a set of personal, household, and community practices are recommended as chain-breaking measures, along with public immunization as the most effective strategy in response to viral pandemics. These include (1) physical and social distancing as general advice and an effective non-pharmaceutical intervention at both the individual and community levels, as manifested by school closures, workplace measures, public transport restrictions, and stay-at-home recommendations [94]; (2) changing social greetings (e.g., handshaking, kissing, and hugging) and face-touching behaviors [95]; (3) personal hygiene by regular handwashing with water and proper detergent like soap (at least 40–60 seconds) or sanitizing one’s hands using alcohol-based sanitizers (62%–71%) for an adequate time if washing is not possible [96]; (4) employing sanitization procedures for commonly shared objects and touched areas by using ultraviolet irradiation or effective disinfectants and chemical compounds to inactivate the landed virus on physical surfaces [96]; and (5) self-protection using personal protective equipment. In this regard, wearing a well-fitted N95, medical, or even homemade mask in compliance with design standards reduces the risk of respiratory emissions, protects against virus exposure, and subsequently from being infected in high-density are where being within close contact with others is difficult to avoid [97].
- For healthcare workers who are at the front line in healthcare settings, there are additional recommendations to take care of themselves and be safe against viral transmission. Although respiratory protection (wearing disposable and well-fitting facial masks [reported to be light, comfortable to wear, and easy to remove] and well-covered facial shields [reported to involve less skin irritation, and easy to breathe through] to block expelled virus-containing respiratory secretions) is highly recommended, eye protection (in addition to other required personal protective equipment) also needs to be carefully considered to limit the risk of viral transmission through the ocular mucous membranes in direct visits and care [98,99]. For this goal, facial shields as an effective barrier may offer a level of protection for the eyes, nose, mouth, and face; however, they might not be welcomed by some workers. Although goggles are not comfortable to wear for hours in daily practice and may interfere with vision, they can be an option to protect the eyes against contamination via droplets and/or splashes [100]. Healthcare workers are also required to wear protective long-sleeved gowns to cover exposed body parts against infected biofluids (i.e., respiratory secretions) and disposable medical gloves (from entry to exit of the patient’s room) to keep their hands clean and avoid virus spread and self-contamination; however, skin irritation (i.e., rash, itching, and dry skin) are expected in some workers [101–103].
- Vaccine, Vaccination, and Herd Immunity
- To end the current viral outbreak, similar to previously reported outbreaks caused by infectious viruses, the development of prophylactic vaccines and subsequently public immunization on a large scale is an urgent priority for all at-risk and affected nations, sub-nations, and territories. Since the start of the recent outbreak, many institutions and companies have been working on both conventional and novel technological innovations using the whole virus or its functional components (e.g., the S protein) to make attenuated and inactivated virus vaccines, viral-vector vaccines, protein subunit vaccines, virus-like particle vaccines, and nucleic acid-based vaccines in a competitive environment, aiming to achieve the most desirable and broadly protective vaccines with all included standards such as high quality, favorable safety, and high efficacy at disease prevention for use in those who are at risk of viral infection [104]. To reach this goal, different SARS-CoV-2 proteins (mostly the viral spike protein) were targeted by recent efforts to find and test the best viral component for vaccine research [104]. As of September 19, 2022, 47 vaccines made by different platforms have been approved and used by 201 countries based on the decisions of national authorities and regulatory agencies; however, these vaccines are still clinically monitored under different trials in different countries to confirm their safety and efficacy (https://covid19.trackvaccines.org/). The most welcomed platforms offering a high grade of protection in vaccine receivers are BNT162b2 (an RNA-based vaccine) by Pfizer–BioNTech (efficacy, 95.0%), mRNA-1273 by Moderna (efficacy, 94.1%), and Sputnik V (adenovirus-based vaccine) by the Gamaleya Institute (efficacy, 91.6%) [105]. The following vaccines have also been tested in trials for use in humans, showing a lesser degree of protection: BBIBP-CorV (inactivated virus vaccine) by Sinopharm (efficacy, 79.34%), Covaxin (whole-virion inactivated vaccine) by Bharat Biotech (efficacy, 81.0%), Ad26.COV2.S (recombinant vaccine/adenovirus serotype 26 [Ad26]) by Johnson & Johnson (efficacy, 72.0%) and AZD1222 (recombinant vaccine) by Oxford/AstraZeneca (efficacy, 70.4%) [105]. For administration, almost all these vaccines are delivered by intramuscular injection into the muscles in 1 to 2 injections based on the type of vaccine and on a certain timetable, with the second injection occurring about 1 month after the first injection in almost all vaccines [105]. Generally, the safety of these vaccines is favorable, and most volunteers who were immunized by these vaccines had no complaints of adverse and serious reactions; however, pain and tenderness, fever, headache, fatigue, and nausea are common in some vaccinated individuals [106]. In general, the storage, handling, and transportation of almost all developed vaccines are challenging; hence, controlled conditions (2°C–8°C for most vaccines for a short period) are essential to maintain the vaccines intact and effective [107,108].
- While mass vaccination programs are strongly recommended in this complicated situation to immunize people against viral infection (both symptomatic and even asymptomatic infections) and to prevent viral transmission, hospitalization, and death, herd immunity may not be reached even after natural infection [109,110]. At this time, there are some challenges ahead that complicate the estimation of the herd immunity threshold. Inequalities in vaccine distribution and coverage are a major concern, since these gaps allow viruses to spread worldwide, especially in low- and middle-income countries with vaccination rates <10% [111,112]. In addition, herd immunity remains difficult to achieve in societies where a large proportion of unvaccinated people have concerns about vaccine safety and necessity [109]. It also should be noted that the immune responses offered by different vaccine platforms are variable in duration, and vaccines differ from each other in terms of efficacy or effectiveness [113]. Additionally, as mentioned in section 2, SARS-CoV-2 has different strains; therefore, infection with one variant may not trigger long-lasting immunity to protect against infection with other variants [108].
Prevention
- After about 2 and a half years of struggles, the current SARS-CoV-2 pandemic is still a matter of concern as the virus tends to continue to genetically evolve into different variants, globally spread with a rapid distribution among human populations, and infect new cases among those who are not immune or do not follow the policies and recommended guidelines on the disease prevention. This virus is not the first emerging human CoV, but it is the most challenging strain, with a high degree of infectiousness and a high interhuman transmission rate. Although dozens of effective vaccines have been designed and are now available to use for public immunization, personal protection by taking appropriate precautions and following relevant guidance of the local health authorities are also essential to stop viral transmission and end the global outbreak. With little knowledge about the behavior of the virus, which seems to be the tip of the iceberg, intensive studies are recommended to comprehensively understand and find answers to open questions regarding the original, epidemiological, pathophysiological, and clinical aspects of SARS-CoV-2 and its associated disease in the near future and to design and develop new and more effective preventive and therapeutic interventions aiming to return the current complicated situation to normal.
Conclusion
-
Ethics Approval
Not applicable.
-
Conflicts of Interest
The authors have no conflicts of interest to declare.
-
Funding
None.
-
Availability of Data
All data generated or analyzed during this study are included in this published article. Other data may be requested from the corresponding author.
-
Authors’ Contributions
Conceptualization: HK; Data curation: HK; Investigation: HK, ZK, NK; Supervision: HK; Writing–original draft: HK; Writing–review & editing: HK, ZK, NK.
-
Additional Contributions
The images that constitute Figures 1–5 were provided by Negin Karami (Alborz University of Medical Sciences, Karaj, Iran).
Article information

Sources: https://cov-lineages.org and https://viralzone.expasy.org.
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; WHO, World Health Organization.
- 1. Zhou Z, Qiu Y, Ge X. The taxonomy, host range and pathogenicity of coronaviruses and other viruses in the Nidovirales order. Anim Dis 2021;1:5. ArticlePubMedPMCPDF
- 2. Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 2016;3:237−61.ArticlePubMedPMC
- 3. Hebbani AV, Pulakuntla S, Pannuru P, et al. COVID-19: comprehensive review on mutations and current vaccines. Arch Microbiol 2021;204:8. ArticlePubMedPMCPDF
- 4. Zhong NS, Zheng BJ, Li YM, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 2003;362:1353−8.ArticlePubMedPMC
- 5. Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012;367:1814−20.ArticlePubMed
- 6. Wiersinga WJ, Rhodes A, Cheng AC, et al. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA 2020;324:782−93.ArticlePubMed
- 7. Ge H, Wang X, Yuan X, et al. The epidemiology and clinical information about COVID-19. Eur J Clin Microbiol Infect Dis 2020;39:1011−9.ArticlePubMedPMCPDF
- 8. Hu B, Guo H, Zhou P, et al. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 2021;19:141−54.ArticlePubMedPDF
- 9. Osuchowski MF, Winkler MS, Skirecki T, et al. The COVID-19 puzzle: deciphering pathophysiology and phenotypes of a new disease entity. Lancet Respir Med 2021;9:622−42.ArticlePubMedPMC
- 10. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382:727−33.ArticlePubMedPMC
- 11. Kumar S, Nyodu R, Maurya VK, et al. Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In: Saxena SK, editor. Coronavirus disease 2019 (COVID-19): epidemiology, pathogenesis, diagnosis, and therapeutics. Singapore: Springer; 2020. p. 23−31.
- 12. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 2015;1282:1−23.ArticlePubMedPMC
- 13. Yadav R, Chaudhary JK, Jain N, et al. Role of structural and non-structural proteins and therapeutic targets of SARS-CoV-2 for COVID-19. Cells 2021;10:821. ArticlePubMedPMC
- 14. Xiang R, Yu Z, Wang Y, et al. Recent advances in developing small-molecule inhibitors against SARS-CoV-2. Acta Pharm Sin B 2022;12:1591−623.ArticlePubMed
- 15. Jaimes JA, Andre NM, Chappie JS, et al. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol 2020;432:3309−25.ArticlePubMedPMC
- 16. Zhou H, Ji J, Chen X, et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell 2021;184:4380−91.ArticlePubMedPMC
- 17. Calistri P, Decaro N, Lorusso A. SARS-CoV-2 pandemic: not the first, not the last. Microorganisms 2021;9:433. ArticlePubMedPMC
- 18. Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020;27:325−8.ArticlePubMedPMC
- 19. Zandi M, Karami H, Soltani S. Role of hemagglutinin esterase protein in neurological manifestation of COVID-19. Fluids Barriers CNS 2021;18:39. ArticlePubMedPMCPDF
- 20. Ciotti M, Ciccozzi M, Pieri M, et al. The COVID-19 pandemic: viral variants and vaccine efficacy. Crit Rev Clin Lab Sci 2022;59:66−75.ArticlePubMed
- 21. Young M, Crook H, Scott J, et al. COVID-19: virology, variants, and vaccines. BMJ Med 2022;1:e000040.ArticlePubMedPMC
- 22. Ramesh S, Govindarajulu M, Parise RS, et al. Emerging SARS-CoV-2 variants: a review of its mutations, its implications and vaccine efficacy. Vaccines (Basel) 2021;9:1195. ArticlePubMedPMC
- 23. Singh J, Pandit P, McArthur AG, et al. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol J 2021;18:166. ArticlePubMedPMCPDF
- 24. Xiao X, Newman C, Buesching CD, et al. Animal sales from Wuhan wet markets immediately prior to the COVID-19 pandemic. Sci Rep 2021;11:11898. ArticlePubMedPMCPDF
- 25. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579:270−3.PubMedPMC
- 26. Liu P, Jiang JZ, Wan XF, et al. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog 2020;16:e1008421.ArticlePubMedPMC
- 27. Andersen KG, Rambaut A, Lipkin WI, et al. The proximal origin of SARS-CoV-2. Nat Med 2020;26:450−2.ArticlePubMedPMCPDF
- 28. Segreto R, Deigin Y. The genetic structure of SARS-CoV-2 does not rule out a laboratory origin: SARS-COV-2 chimeric structure and furin cleavage site might be the result of genetic manipulation. Bioessays 2021;43:e2000240.PubMed
- 29. Tyshkovskiy A, Panchin AY. There is no evidence of SARS-CoV-2 laboratory origin: response to Segreto and Deigin (DOI: 10.1002/bies.202000240). Bioessays 2021;43:e2000325.ArticlePubMedPDF
- 30. Hao P, Zhong W, Song S, et al. Is SARS-CoV-2 originated from laboratory?: a rebuttal to the claim of formation via laboratory recombination. Emerg Microbes Infect 2020;9:545−7.ArticlePubMedPMC
- 31. Horton R. Offline: The origin story-towards a final resolution? Lancet 2022;399:11. ArticlePubMed
- 32. Cevik M, Kuppalli K, Kindrachuk J, et al. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ 2020;371:m3862. ArticlePubMed
- 33. Jayaweera M, Perera H, Gunawardana B, et al. Transmission of COVID-19 virus by droplets and aerosols: a critical review on the unresolved dichotomy. Environ Res 2020;188:109819. ArticlePubMedPMC
- 34. Sosnowski TR. Inhaled aerosols: their role in COVID-19 transmission, including biophysical interactions in the lungs. Curr Opin Colloid Interface Sci 2021;54:101451. ArticlePubMedPMC
- 35. Guo ZD, Wang ZY, Zhang SF, et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg Infect Dis 2020;26:1583−91.ArticlePubMed
- 36. van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med 2020;382:1564−7.ArticlePubMedPDF
- 37. Cevik M, Marcus JL, Buckee C, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission dynamics should inform policy. Clin Infect Dis 2021;73(Suppl 2). S170−6.ArticlePubMedPDF
- 38. Karimzadeh S, Bhopal R, Nguyen Tien H. Review of infective dose, routes of transmission and outcome of COVID-19 caused by the SARS-COV-2: comparison with other respiratory viruses. Epidemiol Infect 2021;149:e96.ArticlePubMed
- 39. Kraay AN, Hayashi MA, Berendes DM, et al. Risk for fomite-mediated transmission of SARS-CoV-2 in child daycares, schools, nursing homes, and offices. Emerg Infect Dis 2021;27:1229−31.ArticlePubMedPMC
- 40. Tharayil A, Rajakumari R, Mozetic M, et al. Contact transmission of SARS-CoV-2 on fomite surfaces: surface survival and risk reduction. Interface Focus 2021;12:20210042. ArticlePubMedPMCPDF
- 41. Rocha AL, Pinheiro JR, Nakamura TC, et al. Fomites and the environment did not have an important role in COVID-19 transmission in a Brazilian mid-sized city. Sci Rep 2021;11:15960. ArticlePubMedPMCPDF
- 42. Chambers C, Krogstad P, Bertrand K, et al. Evaluation for SARS-CoV-2 in breast milk from 18 infected women. JAMA 2020;324:1347−48.ArticlePubMedPMC
- 43. Zamaniyan M, Ebadi A, Aghajanpoor S, et al. Preterm delivery, maternal death, and vertical transmission in a pregnant woman with COVID-19 infection. Prenat Diagn 2020;40:1759−61.ArticlePubMedPMCPDF
- 44. Andersson MI, Arancibia-Carcamo CV, Auckland K, et al. SARS-CoV-2 RNA detected in blood products from patients with COVID-19 is not associated with infectious virus. Wellcome Open Res 2020;5:181. ArticlePubMedPMCPDF
- 45. Li D, Jin M, Bao P, et al. Clinical characteristics and results of semen tests among men with coronavirus disease 2019. JAMA Netw Open 2020;3:e208292.ArticlePubMedPMC
- 46. Jackson CB, Farzan M, Chen B, et al. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 2022;23:3−20.ArticlePubMedPDF
- 47. Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020;181:281−92.ArticlePubMedPMC
- 48. Liu J, Li Y, Liu Q, et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov 2021;7:17. ArticlePubMedPMCPDF
- 49. Lanjanian H, Moazzam-Jazi M, Hedayati M, et al. SARS-CoV-2 infection susceptibility influenced by ACE2 genetic polymorphisms: insights from Tehran Cardio-Metabolic Genetic Study. Sci Rep 2021;11:1529. ArticlePubMedPMCPDF
- 50. Gu Y, Cao J, Zhang X, et al. Receptome profiling identifies KREMEN1 and ASGR1 as alternative functional receptors of SARS-CoV-2. Cell Res 2022;32:24−37.ArticlePubMedPDF
- 51. Shafqat A, Shafqat S, Salameh SA, et al. Mechanistic insights into the immune pathophysiology of COVID-19; an in-depth review. Front Immunol 2022;13:835104. ArticlePubMedPMC
- 52. Rahimi B, Vesal A, Edalatifard M. Coronavirus and Its effect on the respiratory system: is there any association between pneumonia and immune cells. J Family Med Prim Care 2020;9:4729−35.ArticlePubMedPMC
- 53. Ahn JH, Kim J, Hong SP, et al. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J Clin Invest 2021;131:e148517.ArticlePubMedPMC
- 54. Drozdzik A, Drozdzik M. Oral pathology in COVID-19 and SARS-CoV-2 infection-molecular aspects. Int J Mol Sci 2022;23:1431. ArticlePubMedPMC
- 55. Huang N, Perez P, Kato T, et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat Med 2021;27:892−903.PubMedPMC
- 56. Lamers MM, Haagmans BL. SARS-CoV-2 pathogenesis. Nat Rev Microbiol 2022;20:270−84.ArticlePubMedPDF
- 57. Shahgolzari M, Yavari A, Arjeini Y, et al. Immunopathology and Immunopathogenesis of COVID-19, what we know and what we should learn. Gene Rep 2021;25:101417. ArticlePubMedPMC
- 58. Beyer DK, Forero A. Mechanisms of antiviral immune evasion of SARS-CoV-2. J Mol Biol 2022;434:167265. ArticlePubMed
- 59. Tay MZ, Poh CM, Renia L, et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol 2020;20:363−74.ArticlePubMedPMCPDF
- 60. Marik PE, Iglesias J, Varon J, et al. A scoping review of the pathophysiology of COVID-19. Int J Immunopathol Pharmacol 2021;35:20587384211048026. ArticlePubMedPMCPDF
- 61. Quan C, Li C, Ma H, et al. Immunopathogenesis of coronavirus-induced acute respiratory distress syndrome (ARDS): potential infection-associated hemophagocytic lymphohistiocytosis. Clin Microbiol Rev 2020;34:e00074−20.ArticlePubMedPMCPDF
- 62. Varghese PM, Tsolaki AG, Yasmin H, et al. Host-pathogen interaction in COVID-19: pathogenesis, potential therapeutics and vaccination strategies. Immunobiology 2020;225:152008. ArticlePubMedPMC
- 63. Vardhana SA, Wolchok JD. The many faces of the anti-COVID immune response. J Exp Med 2020;217:e20200678.ArticlePubMedPMCPDF
- 64. Bhardwaj A, Sapra L, Saini C, et al. COVID-19: immunology, immunopathogenesis and potential therapies. Int Rev Immunol 2022;41:171−206.ArticlePubMed
- 65. Wu ML, Liu FL, Sun J, et al. SARS-CoV-2-triggered mast cell rapid degranulation induces alveolar epithelial inflammation and lung injury. Signal Transduct Target Ther 2021;6:428. ArticlePubMedPMCPDF
- 66. Schousboe P, Ronit A, Nielsen HB, et al. Reduced levels of pulmonary surfactant in COVID-19 ARDS. Sci Rep 2022;12:4040. ArticlePubMedPMCPDF
- 67. Diao B, Wang C, Tan Y, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol 2020;11:827. ArticlePubMedPMC
- 68. Gupta A, Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID-19. Nat Med 2020;26:1017−32.ArticlePubMedPDF
- 69. Jin JM, Bai P, He W, et al. Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health 2020;8:152. ArticlePubMedPMC
- 70. Dabisch PA, Biryukov J, Beck K, et al. Seroconversion and fever are dose-dependent in a nonhuman primate model of inhalational COVID-19. PLoS Pathog 2021;17:e1009865.ArticlePubMedPMC
- 71. Rahman HS, Abdulateef DS, Hussen NH, et al. Recent advancements on COVID-19: a comprehensive review. Int J Gen Med 2021;14:10351−72.ArticlePubMedPMCPDF
- 72. Stokes EK, Zambrano LD, Anderson KN, et al. Coronavirus disease 2019 case surveillance: United States, January 22-May 30, 2020. MMWR Morb Mortal Wkly Rep 2020;69:759−65.ArticlePubMedPMC
- 73. Bajgain KT, Badal S, Bajgain BB, et al. Prevalence of comorbidities among individuals with COVID-19: a rapid review of current literature. Am J Infect Control 2021;49:238−46.ArticlePubMed
- 74. Tsai PH, Lai WY, Lin YY, et al. Clinical manifestation and disease progression in COVID-19 infection. J Chin Med Assoc 2021;84:3−8.ArticlePubMed
- 75. Islam MA, Kundu S, Alam SS, et al. Prevalence and characteristics of fever in adult and paediatric patients with coronavirus disease 2019 (COVID-19): a systematic review and meta-analysis of 17515 patients. PLoS One 2021;16:e0249788.ArticlePubMedPMC
- 76. Ng DH, Choy CY, Chan YH, et al. Fever patterns, cytokine profiles, and outcomes in COVID-19. Open Forum Infect Dis 2020;7:ofaa375. ArticlePubMedPMCPDF
- 77. Gul MH, Htun ZM, Inayat A. Role of fever and ambient temperature in COVID-19. Expert Rev Respir Med 2021;15:171−3.ArticlePubMed
- 78. Grudlewska-Buda K, Wiktorczyk-Kapischke N, Walecka-Zacharska E, et al. SARS-CoV-2-morphology, transmission and diagnosis during pandemic, review with element of meta-analysis. J Clin Med 2021;10:1962. ArticlePubMedPMC
- 79. Bertolino L, Vitrone M, Durante-Mangoni E. Does this patient have COVID-19? A practical guide for the internist. Intern Emerg Med 2020;15:791−800.ArticlePubMedPMCPDF
- 80. Parasher A. COVID-19: current understanding of its pathophysiology, clinical presentation and treatment. Postgrad Med J 2021;97:312−20.ArticlePubMed
- 81. Yong SJ. Long COVID or post-COVID-19 syndrome: putative pathophysiology, risk factors, and treatments. Infect Dis (Lond) 2021;53:737−54.ArticlePubMed
- 82. Lopez-Leon S, Wegman-Ostrosky T, Perelman C, et al. More than 50 long-term effects of COVID-19: a systematic review and meta-analysis. Sci Rep 2021;11:16144. ArticlePubMedPMCPDF
- 83. Castanares Zapatero D, Hanquet G, Van den Heede K. Epidemiology of long COVID: a pragmatic review of the literature. Brussels: KCE; 2021.
- 84. Bai F, Tomasoni D, Falcinella C, et al. Female gender is associated with long COVID syndrome: a prospective cohort study. Clin Microbiol Infect 2022;28:611. Article
- 85. Sudre CH, Murray B, Varsavsky T, et al. Attributes and predictors of long COVID. Nat Med 2021;27:626−31.ArticlePubMedPMCPDF
- 86. Subramanian A, Nirantharakumar K, Hughes S, et al. Symptoms and risk factors for long COVID in non-hospitalized adults. Nat Med 2022;28:1706−14.ArticlePubMedPMCPDF
- 87. Thompson EJ, Williams DM, Walker AJ, et al. Long COVID burden and risk factors in 10 UK longitudinal studies and electronic health records. Nat Commun 2022;13:3528. ArticlePubMedPMCPDF
- 88. Asadi-Pooya AA, Akbari A, Emami A, et al. Risk factors associated with long COVID syndrome: a retrospective study. Iran J Med Sci 2021;46:428−36.PubMedPMC
- 89. Arjun MC, Singh AK, Pal D, et al. Prevalence, characteristics, and predictors of long COVID among diagnosed cases of COVID-19 [Preprint]. Posted 2022 Jan 8. medRxiv 2022.01.04.21268536. https://doi.org/10.1101/2022.01.04.21268536.Article
- 90. Aiyegbusi OL, Hughes SE, Turner G, et al. Symptoms, complications and management of long COVID: a review. J R Soc Med 2021;114:428−42.ArticlePubMedPMCPDF
- 91. Mehandru S, Merad M. Pathological sequelae of long-haul COVID. Nat Immunol 2022;23:194−202.ArticlePubMedPMCPDF
- 92. Ramakrishnan RK, Kashour T, Hamid Q, et al. Unraveling the mystery surrounding post-acute sequelae of COVID-19. Front Immunol 2021;12:686029. ArticlePubMedPMC
- 93. Peluso MJ, Deeks SG. Early clues regarding the pathogenesis of long-COVID. Trends Immunol 2022;43:268−70.ArticlePubMedPMC
- 94. Sun KS, Lau TS, Yeoh EK, et al. Effectiveness of different types and levels of social distancing measures: a scoping review of global evidence from earlier stage of COVID-19 pandemic. BMJ Open 2022;12:e053938.ArticlePubMed
- 95. Ayre J, Cvejic E, McCaffery K, et al. Contextualising COVID-19 prevention behaviour over time in Australia: patterns and long-term predictors from April to July 2020 in an online social media sample. PLoS One 2021;16:e0253930.ArticlePubMedPMC
- 96. Cirrincione L, Plescia F, Ledda C, et al. COVID-19 pandemic: prevention and protection measures to be adopted at the workplace. Sustainability 2020;12:3603. Article
- 97. Liao M, Liu H, Wang X, et al. A technical review of face mask wearing in preventing respiratory COVID-19 transmission. Curr Opin Colloid Interface Sci 2021;52:101417. ArticlePubMedPMC
- 98. Alzunitan MA, Perencevich EN, Edmond MB. Assessing health care worker perceptions of face coverings during the COVID-19 pandemic. Am J Infect Control 2021;49:521−2.ArticlePubMed
- 99. Byambasuren O, Beller E, Clark J, et al. The effect of eye protection on SARS-CoV-2 transmission: a systematic review. Antimicrob Resist Infect Control 2021;10:156. ArticlePubMedPMCPDF
- 100. Xiao-huan H, Yan-ru F, Gao-ming L, et al. The impact of goggle-associated harms to health and working status of nurses during management of COVID-19 [Preprint]. Posted 2020 May 15. medRxiv 2020.05.11.20094854. https://doi.org/10.1101/2020.05.11.20094854.Article
- 101. Aumeran C, Henquell C, Brebion A, et al. Isolation gown contamination during healthcare of confirmed SARS-CoV-2-infected patients. J Hosp Infect 2021;107:111−3.ArticlePubMed
- 102. Huang F, Armando M, Dufau S, et al. COVID-19 outbreak and healthcare worker behavioural change toward hand hygiene practices. J Hosp Infect 2021;111:27−34.ArticlePubMedPMC
- 103. Hu K, Fan J, Li X, et al. The adverse skin reactions of health care workers using personal protective equipment for COVID-19. Medicine (Baltimore) 2020;99:e20603.ArticlePubMedPMC
- 104. Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol 2021;21:73−82.ArticlePubMedPMCPDF
- 105. Kantarcioglu B, Iqbal O, Walenga JM, et al. An update on the pathogenesis of COVID-19 and the reportedly rare thrombotic events following vaccination. Clin Appl Thromb Hemost 2021;27:10760296211021498. ArticlePubMedPMCPDF
- 106. Hadj Hassine I. COVID-19 vaccines and variants of concern: a review. Rev Med Virol 2022;32:e2313.PubMed
- 107. Rogers B, Dennison K, Adepoju N, et al. Vaccine cold chain: part 1. proper handling and storage of vaccine. AAOHN J 2010;58:337−4.ArticlePubMedPDF
- 108. Torres-Estrella CU, Reyes-Montes MD, Duarte-Escalante E, et al. Vaccines against COVID-19: a review. Vaccines (Basel) 2022;10:414. ArticlePubMedPMC
- 109. Aschwanden C. Five reasons why COVID herd immunity is probably impossible. Nature 2021;591:520−2.ArticlePubMedPDF
- 110. Dye C. The benefits of large scale COVID-19 vaccination. BMJ 2022;377:o867. ArticlePubMed
- 111. Rydland HT, Friedman J, Stringhini S, et al. The radically unequal distribution of COVID-19 vaccinations: a predictable yet avoidable symptom of the fundamental causes of inequality. Humanit Soc Sci Commun 2022;9:1−6.ArticlePDF
- 112. Pilkington V, Keestra SM, Hill A. Global COVID-19 vaccine inequity: failures in the first year of distribution and potential solutions for the future. Front Public Health 2022;10:821117. ArticlePubMedPMC
- 113. Fiolet T, Kherabi Y, MacDonald CJ, et al. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: a narrative review. Clin Microbiol Infect 2022;28:202−21.ArticlePubMedPMC
References
Figure & Data
References
Citations

- Polysaccharides and Lectins: A Natural Complementary Approach against the SARS-CoV-2 Pandemic
Radu Lefter, Prairna Balyan, Ioana-Miruna Balmus, Abdellah Ech-Chahad, Ahmad Ali, Alin Ciobica, Antoneta Dacia Petroaie, Gabriela Halitchi, Bogdan Novac, Catalina Ionescu, Fatima Zahra Kamal
Microbiology Research.2024; 15(2): 525. CrossRef - Surveillance of endemic coronaviruses during the COVID‐19 pandemic in Iran, 2021–2022
Hassan Karami, Kaveh Sadeghi, Sevrin Zadheidar, Fatemeh Saadatmand, Negar Mirsalehi, Nima Hoveidi Ardestani, Shirin Kalantari, Mohammad Farahmand, Jila Yavarian, Talat Mokhtari‐Azad
Influenza and Other Respiratory Viruses.2023;[Epub] CrossRef


 Cite
Cite