Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 5(6); 2014 > Article
-
Original Article
Diversity of Rotavirus Strain Circulated in Gwangju, Republic of Korea - Min Ji Kima, Hye Sook Jeongb, Seon Gyeong Kima, Se Mi Leea, Sun Hee Kima, Hye-Young Keea, Eun-hye Joa, Hye-jung Parka, Dong-Ryong Haa, Eun Sun Kima, Kye-Won Seoa, Jae Keun Chunga
-
Osong Public Health and Research Perspectives 2014;5(6):364-369.
DOI: https://doi.org/10.1016/j.phrp.2014.10.004
Published online: November 1, 2014
aMicrobiology Division, Health and Environment Research Institute of Gwangju, Gwangju, Korea
bDivision of Vaccine Research, Korea National Institute of Health, Cheongju, Korea
- ∗Corresponding author. jkchung@korea.kr
• Received: August 14, 2014 • Revised: September 18, 2014 • Accepted: October 1, 2014
© 2014 Published by Elsevier B.V. on behalf of Korea Centers for Disease Control and Prevention.
This is an Open Access article distributed under the terms of the CC-BY-NC License (http://creativecommons.org/licenses/by-nc/3.0).
Abstract
-
Objectives
- The introduction of new rotavirus vaccines into the public sphere makes it necessary to maintain constant surveillance and to heighten public awareness of the appearance of new rotavirus strains. We describe the molecular epidemiology of circulating rotavirus strains after vaccine introduction.
-
Methods
- We collected a total of 1070 stool samples from children with gastroenteritis from January 2013 to June 2013. The antigenic prevalence of rotavirus group A was distinguished using enzyme immunoassay. The G and P genotypes of enzyme immunoassay-positive samples were determined with reverse transcription-polymerase chain reaction and nucleotide sequencing analysis.
-
Results
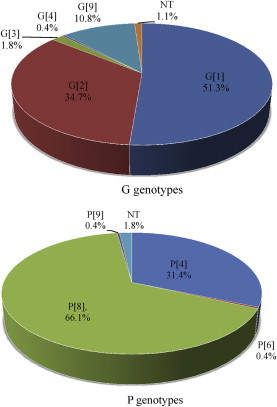
- Of the 1070 samples collected, 277 (25.9%) tested positive for rotaviruses by enzyme-linked immunoabsorbent assay. The most prevalent circulating genotype G was G1 (51.3%), followed by G2 (34.7%) and G9 (10.8%). The predominant type of genotype P was P[8] (66.1%), followed by P[4] (31.4%). In this study, nine genotypes were found. G1P[8] was the most prevalent (51.8%), followed by G2P[4] (30.5%), G9P[8] (9.9%), and G2P[8] (4.0%). Several unusual combinations (G1P[4], G3P[9], G3P[8], G4P[6], and G9P[4]) were also identified.
-
Conclusion
- Molecular epidemiological knowledge of rotaviruses is critical for the development of effective preventive measures, including vaccines. These data will help us monitor the effectiveness of current rotavirus vaccines.
- Rotaviruses (RVs) are the leading cause of acute and severe gastroenteritis, diarrhea, and malnutrition primarily in children younger than 5 years [1], and are responsible for approximately 600,000 deaths worldwide each year [2,3].
- RVs belong to the family Reoviridae. The viral genome consists of 11 segments of double-stranded (ds) RNA genome that encodes six structural proteins (VP1–4, VP6, and VP7) and six nonstructural proteins (NSP1–6) [1]. So far, RVs are subdivided into eight groups (A–H) on the basis of the antigenic properties or the amino acid sequences of the inner capsid protein VP6 [4].
- Based on the differences of VP7 and VP4 gene sequences, RVs are divided into genotypes G and P, respectively. To date, at least 27 G and 35 P genotypes have been reported from humans and a variety of mammalian and avian species [5]. Of these, 11 G genotypes and 12 P genotypes have been isolated from humans [6–8]. However, the most commonly isolated human RV genotypes are a small number of combined G/P genotypes, such as G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8] [8,9]. Certain G and P types have been found to be highly prevalent in different areas around the world such as G5 types in Brazil and G10 types in India [10,11]. More recently, a surveillance program directed by the World Health Organization noted that in 2010 the predominant uncommon strains were G12P[8] and G12P[6] viruses in Southeast Asia; G2P[6], G3P[6], and G1P[6] viruses in sub-Saharan Africa; G1P[4] and G2P[8] viruses in the Western Pacific; and G9P[4] viruses in the Americas [9].
- In South Korea, molecular epidemiological studies have shown that G2, G4, and G9 were the most isolated genotypes from 1998 to 2004. G1 was the most predominant genotype from years 1987 to 1999, and genotypes G2, G3, and G4 were also isolated during the same period [12]. G11, a rare strain, was also reported for the first time in South Korea in 2007 [13]. Recently, there have been changes in the frequency of genotype isolations from more common G/P combinations with the exceptions of G1P[8] and G3P[8] to the rare combinations of genotypes such as G4P[6] and G2P[8] [14]. These distinct changes in the prevalence of circulating RV strains suggest that surveillance studies are important for the successful vaccine development and efficacy testing.
- Even with the ongoing vaccinations against RVs, RV infection with high morbidity and mortality occurs. This indicates that we should take into account the role of RV vaccines in the natural temporal variability in genotypes G and P. Two live attenuated RV vaccines were licensed and became commercially available in South Korea. RotaTeq (Merck & Company, Inc, Whitehouse Station, NJ, USA), launched in September 2007, is a vaccine that consists of five distinct bovine reassortants, and each of the five vaccine strains contains outer capsid proteins from a serotype of the human RVs (G1, G2, G3, G4, and P[8]). Rotarix (GlaxoSmithK-line, Rixensart, Belgium), licensed in June 2008, consists of a single attenuated G1P[8] strain of human RV.
- The effect of RV vaccines on the natural pattern of circulating RV strains in human populations is unknown and difficult to predict. Continuing surveillance is needed to identify the spectrum of protection engendered by each vaccine and the effect that each vaccine may have on circulating strains. Documentation of long-term temporal changes in RV strain distributions requires a detailed analysis of targeted monotypes in circulation prior to and after vaccine introduction.
- In this study, we report the distribution of RV genotypes G and P that have been circulated during the first half of 2013 in Gwangju, South Korea.
Introduction
- 2.1 Sample collection
- From January 2013 to June 2013, a total of 1070 stool samples were collected from children who were hospitalized with acute gastroenteritis symptoms in eight hospitals, Gwangju, South Korea. Samples were kept at 4°C until they were transported to the laboratory for analysis. Clinical information on age and sex of patients, dates of disease onset and sample collection, symptoms, RV vaccination history, etc., was recorded as background data.
- 2.2 RV antigen detection
- RV antigens were detected in stool supernatants using enzyme-linked immunoabsorbent assay (ELISA) with VP6 group-specific antibody (BioTracer Rotavirus ELISA kit; Biofocus, Uiwang, Korea) according to the manufacturer's instructions. Specimens with Optic Density (OD) absorbance values >0.4 at a 450-nm wavelength were considered to be positive.
- 2.3 Reverse transcription-polymerase chain reaction for genotyping
- G and P genotyping was performed using reverse transcription-polymerase chain reaction (RT-PCR) on 277 RV ELISA positive samples. Fecal specimens were diluted 1:10 in phosphate-buffered saline. After a thorough mixing, each fecal suspension was centrifuged for 20 minutes at 1000 × g, 4°C.
- RV ds RNA was extracted from 140 μL of 10% fecal suspensions using an RNA extraction Kit [QIAamp Viral RNA mini Kit (spin protocol); Qiagen, Inc., Hilden, Germany] in accordance with the manufacturer's instructions. The extracted RNA was denaturated at 95°C for 5 minutes. RT-PCR was performed for RV G and P genotypes using Accupower Hotstart RT/PCR premix kit (Bioneer, Daejeon, Korea). We amplified an 881-bp fragment of the VP7 gene with the consensus forward primer VP7-F (5′-ATG TAT GGT ATT GAA TAT ACC AC-3′) and reverse primer VP7-R (5′-AAC TTG CCA CCA TTT TTT CC-3′) [15]. We also amplified an 876-bp fragment of the VP4 gene with the consensus forward primer Con3 (5′-TGG CTT CGC CAT TTT ATA GAC A-3′) and the reverse primer Con2 (5′-ATT TCG GAC CAT TTA TAA CC-3′) [16]. The PCR reaction for the VP7 and VP4 gene amplification was carried out with an initial RT step at 42°C for 40 minutes, followed by PCR activation at 94°C for 15 minutes, then 35 cycles of amplification (1 minute at 94°C, 1 minute at 50°C, and 1 minute at 72°C), and a final extension of 10 minutes at 72°C in GeneAmp PCR system 9700 (Applied Biosystems, Boston, MA, USA). The PCR products were then electrophoresed on 1.5% agarose gel.
- 2.4 Nucleotide sequencing and sequence analysis
- PCR products were purified using QIAquick Gel Extraction kit (Qiagen GmbH, Hilden, Germany) in accordance with the manufacturer's instructions. The cycle sequencing reaction was carried out with the ABI PRISM BigDye terminator cycle sequencing reaction kit (Applied Biosystems, Foster City, CA, USA), and sequence data were collected by means of an automated DNA analyzer Applied Biosystems (ABI) Prism 3100 Genetic Analyzer (Life Technologies, Foster City, California, USA). Phylogenetic analysis was performed using the Clustal W algorithm in MEGA 5.0 software package [17], using the Maximum Likelihood method for phylogenetic analysis, with 1000 bootstrap replicates.
Materials and methods
- 3.1 Frequency of RV infection by vaccination status
- Overall, 774 of 1070 patient records clearly stated the status of the vaccination, and the previously vaccinated patients were vaccinated either with RotaTeq or with Rotarix. Sixty-seven (17.4%) of the 385 vaccinated patients were infected with RVs, compared to the infection of 144 (37%) of 389 patients who did not receive the vaccine (Table 1). Statistical significance in two tests was assessed. A p value of <0.05 was considered indicative of a statistically significant difference. All analyses were performed using IBM SPSS statistics 20 (IBM Korea, Seoul, Korea).
- 3.2 Frequencies of G and P genotypes
- RVs were detected in 277 of 1070 (25.9%) collected samples. Two hundred of 277 (86.6%) patients with RV gastroenteritis were older than 5 years. The G genotype was identified for 274 samples (98.8%), and the P genotype was identified for 272 (98.2%) RV samples. The most prevalently circulating G genotype was G1 (51.3%, n = 142), followed by G2 (34.7%, n = 96), G9 (10.8%, n = 30), G3 (1.8%, n = 8), and G4 (0.4%, n = 1). The predominant type of P genotype was P[8] (66.1%, n = 183), which was followed by P[4] (31.4%, n = 87), P[6], and P[9] (0.4%, n = 1) (Figure 1, Table 2).
- 3.3 Distribution of G and P genotype combinations
- In total, 272 G and P combinations made up with nine different genotypes were identified. G1P[8], the most frequently detected strain, was responsible for 51.8% (n = 141) of infections. G2P[4], G9P[8], and G2P[8] were detected with a prevalence rate of 30.5% (n = 83), 9.9% (n = 27), and 4.0% (n = 11), respectively. In addition, unusual RV strains were identified, which bore the genotypes of G1P[4], G3P[9], G3P[8], G4P[6], and G9P[4] (Table 3). Among the 67 patients who were vaccinated with RV, the predominant strain was G2P[4] (40.3%), followed by G1P[8] (39.0%) and G9P[8] (12.5%). And among the 389 who were not vaccinated, 144 rotavirus positive samples were analyzed, and the frequencies were as follows: G1P[8] (56.3%), G2P[4] (28.2%), and G9P[8] (9.86%), which were identified with the indicated prevalence (Figure 2).
Results
- RVs are composed of genetically diverse populations of segmented ds RNA viruses, which cause gastroenteritis in many animal species including humans. From year to year and from place to place, the prevalence of human RV G and P types associated with the disease fluctuates [18].
- In the present study, the distribution of the genotype of Group A RVs in the cases of acute gastroenteritis in Gwangju, South Korea, was investigated to determine the efficacy of currently used RV vaccines in South Korea.
- In our study, 1070 fecal specimens were collected from children hospitalized with diarrhea, and 277 (25.9%) were positive for RV. A nationwide surveillance of RV infection conducted by 16 laboratories of local public health institutes revealed the infection rate of RVs in South Korea from 2008 to 2013. The detection rate of RVs was 8.1% (9048 of 112,344) in Korea from 2008 to 2011, 5.6% (1057 of 18,731) in 2012, and 7.6% (1430 of 18,908) in 2013 [19]. The RV detection rate in Gwangju was higher than the average of the national RV infection rate; it was recorded as 15.1% (1822 of 12,099) in the Gwangju area from 2008 to 2011, 7.2% (160 of 2212) in 2012, and 25.9% (277 of 1070) in 2013. Although RV vaccination was active, the number of patients with rotaviral gastroenteritis in 2013 increased by more than three times when compared with the previous year in Gwangju, South Korea.
- One of the main goals of this study was to characterize the VP7 (G genotype) and VP4 (P genotype) genes from RVs circulating in public. The predominant G genotype was G1 (51.3%), followed by G2 (34.7%) and G9 (10.8%). G2 and G4 RVs were the most frequently detected genotypes out of the four common strains (G1–G4) between 1998 and 2004 [20]. G1 was the most predominant strain for a 10-year period prior to 1997 and during 2005–2009 in South Korea [14]. Recently, human G9 strains became the fifth most common strain found in the public sphere [21]. The first G9 strain, WI91, was detected in children in the United States in 1983 [22] and was first detected in 2002 in South Korea [14]. G9 has become recognized as one of the emerging genotypes in many countries such as Cuba [23], France [24], Mexico [25], the United States [26], and Libya [27]. In this study, G9s were the third most identified genotype and had a slightly greater predominance over G3 and G4. These results suggest that the genotype G9 should be considered one of the serotypes of choice for RV vaccines in the future. P[8] (66.1%) was the most common genotype P, followed by P[4] (31.4%), P[6], and P[9] (0.1%). This result is in agreement with previous data stating that P[8] was the most prevalent strain in Korea and other countries [12,28]. Genotypes P[6] and P[9] were detected less frequently; however, these uncommon types continue to be of interest because of a possible agent in RV vaccine programs.
- Regarding G–P combinations, the G1P[8] (51.8%) genotype combination was the most prevalent RV type followed by G2P[4] (30.5%) in 2013. Previous studies on RV circulation in South Korea from 2005 until 2007 have shown that G1P[8] (36%) was the predominant type, followed by G2P[8] (16%), G4P[6] (8.9%), and G1P[6] (8.2%) [14]. The prevalence of G2P[4] was 34.6% in 2000–2001 [29]. Recently, an increase in the incidence of infection with the G2P[4] strain has been reported in Brazil, where the monovalent G1P[8] vaccine is currently in use [30]. Also, reassortants of common human strains, G9P[4] and G1P[4], and a zoonotic genotype G4P[6] were also identified in south Korea [29].
- These studies suggest that the distribution of circulating RV genotypes and predominant strains changes from year to year and has been reported to differ in different regions. Moreover, there are reassortments between human RVs (P[8] and P[9]) or between human and animal strains. Therefore, continuing surveillance is important to monitor the changes in virus genotype patterns and the appearance of new genotypes in each country.
- There were slight differences in the genotype distribution in the patient groups with or without prior vaccination. We conclude that the prevalence of specific RV genotypes in a region changes each year, and nonvaccine strains such as G9, P[4], P[6], and P[9] have increased in Gwangju, South Korea. These emerging strains should be considered as candidates for new RV vaccines in the future.
Discussion
- The authors declare no conflicts of interest.
Conflicts of interest
-
Acknowledgements
- This study was supported by the laboratory surveillance system for acute gastroenteritis in Korea from the National Institute of Health, Korea Centers for Disease Control and Prevention.
Acknowledgments
-
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Article information
- 1. Estes M.K., Kapikian A.Z.. Rotaviruses. Edited by Knipe D.M., Howley P.M., Griffin D.E.: Fields virology. 5th ed.2007. Lippincott Williams and Wilkins; Philadelphia, PA: pp 1917−1974.
- 2. Parashar U.D., Gibson C.J., Bresee J.S.. Rotavirus and severe childhood diarrhoea. Emerg Infect Dis 12(2). 2006 Feb;304−306. PMID: 16494759.ArticlePubMed
- 3. WHO . Rotavirus vaccines. WHO position paper. Wkly Epidemiol Rec 82:2007 Aug;285−296. PMID: 17691162.PubMed
- 4. Matthijnssens J., Otto P.H., Ciarlet M.. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch Virol 157(6). 2012 Jun;1177−1182. PMID: 22430951.ArticlePubMed
- 5. Matthijnssens J., Ciarlet M., McDonald S.M.. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 156(8). 2011 Aug;1397−1413. PMID: 21597953.ArticlePubMed
- 6. Matthijnssens J., Joelsson D.B., Warakomski D.J.. Molecular and biological characterization of the 5 human–bovine rotavirus (WC3)-based reassortant strains of the pentavalent rotavirus vaccine, RotaTeq. Virology 403(2). 2010 Aug;111−127. PMID: 20451234.ArticlePubMed
- 7. Esona M.D., Steele D., Kerin T.. Determination of the G and P types of previously nontypeable rotavirus strains from the African Rotavirus Network, 1996–2004: identification of unusual G types. J Infect Dis 202(Suppl. 1). 2010;S49−54. PMID: 20684717.ArticlePubMed
- 8. Santos N., Hoshino Y.. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol 15(1). 2005 Jan–Feb;29−56. PMID: 15484186.ArticlePubMed
- 9. WHO . Global rotavirus information and surveillance bulletin. Reporting period: January through December 2010. vol. 4:2011. World Health Organization.
- 10. Gentsch J.R., Woods P.A., Ramachandran M.. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J Infect Dis 174(Suppl. 1). 1996;S30−S36. PMID: 8752288.ArticlePubMed
- 11. Ramachandran M., Das B.K., Vij A.. Unusual diversity of human rotavirus G and P genotypes in India. J Clin Microbiol 34(2). 1996 Feb;436−439. PMID: 8789033.ArticlePubMed
- 12. Le V.P., Kim J.Y., Cho S.L.. Detection of unusual rotavirus genotypes G8P[8] and G12P[6] in South Korea. J Med Virol 80(1). 2008 Jan;175−182. PMID: 18041003.ArticlePubMed
- 13. Hong S.K., Lee S.G., Lee S.A.. Characterization of a G11, P [4] strain of human rotavirus isolated in South Korea. J Clin Microbiol 45(11). 2007 Nov;3759−3761. PMID: 17728473.ArticlePubMed
- 14. Lee S.Y., Hong S.K., Lee C.I.. Human rotavirus genotypes in hospitalized children, South Korea, April 2005 to March 2007. Vaccine 27(5). 2009 Nov;97−101.Article
- 15. Iturriza-Gómara M., Cubitt D., Desselberger U.. Amino acid substitution within the VP7 protein of G2 rotavirus strains associated with failure to serotype. J Clin Microbiol 39(10). 2001 Oct;3796−3798. PMID: 11574622.ArticlePubMed
- 16. Gentsch J.R., Glass R.I., Woods P.. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol 30(6). 1992 Jun;1365−1373. PMID: 1320625.ArticlePubMed
- 17. Tamura K., Peterson D., Peterson N.. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10). 2011;2731−2739. PMID: 21546353.ArticlePubMedPMC
- 18. Patton J.T.. Rotavirus diversity and evolution in the post-vaccine world. Discov Med 13(68). 2012 Jan;85−97. PMID: 22284787.PubMed
- 19.
- 20. Song M.O., Kim K.J., Chung S.I.. Distribution of human group a rotavirus VP7 and VP4 types circulating in Seoul, Korea between 1998 and 2000. J Med Virol 70(2). 2003 Jun;324−328. PMID: 12696125.ArticlePubMed
- 21. Kim J.S., Kang J.O., Cho S.C.. Epidemiological profile of rotavirus infection in the Republic of Korea: results from prospective surveillance in the Jeongeub District, 1 July 2002 through 30 June 2004. J Infect Dis 192(Suppl. 1). 2005;S49−56. PMID: 16088805.ArticlePubMed
- 22. Clark H.F., Hoshino Y., Bell L.M.. Rotavirus isolate WI61 representing a presumptive new human serotype. J Clin Microbiol 25(9). 1987 Sep;1757−1762. PMID: 2443534.ArticlePubMed
- 23. Ribas M., Nagashima S., Calzado A.. Emergence of G9 as a predominant genotype of human rotaviruses in Cuba. J Med Virol 83(4). 2011 Apr;738−744. PMID: 21328392.ArticlePubMed
- 24. de Rougemont A., Kaplon J., Pillet S.. Molecular and clinical characterization of rotavirus from diarrheal infants admitted to pediatric emergency units in France. Pediatr Infect Dis J 30(2). 2011 Feb;118−124. PMID: 20686439.ArticlePubMed
- 25. Yen C., Figueroa J.R., Uribe E.S.. Monovalent rotavirus vaccine provides protection against an emerging fully heterotypic G9P[4] rotavirus strain in Mexico. J Infect Dis 204(5). 2011 Sep 1;783−786. PMID: 21810915.Article
- 26. Abdel-Haq N., Amjad M., McGrath E.. Emergence of human rotavirus genotype G9 in metropolitan Detroit between 2007 and 2009. J Med Microbiol 60(Pt 6). 2011 Jun;761−767. PMID: 21372186.ArticlePubMed
- 27. Abugalia M., Cuevas L., Kirby A.. Clinical features and molecular epidemiology of rotavirus and norovirus infections in Libyan children. J Med Virol 83(10). 2011 Oct;1849−1856. PMID: 21837804.ArticlePubMed
- 28. Lennon G., Reidy N., Cryan B.. Changing profile of rotavirus in Ireland: predominance of P[8] and emergence of P[6] and P[9] in mixed infections. J Med Virol 80(3). 2008 Mar;524−530. PMID: 18205218.ArticlePubMed
- 29. Jeong H.S., Lee K.B., Jeong A.Y.. Genotypes of the circulating rotavirus strains in the seven prevaccine seasons from September 2000 to August 2007 in South Korea. Clin Microbiol Infect 17(2). 2011 Feb;232−235. PMID: 20384698.ArticlePubMed
- 30. Gurgel R.Q., Cuevas L.E., Vieira S.C.. Predominance of rotavirus P[4]G2 in a vaccinated population, Brazil. Emerg Infect Dis 13(10). 2007;1571−1573. PMID: 18258011.ArticlePubMedPMC
References
Figure 2Distribution of major genotypes of rotavirus in Gwangju, with respect to vaccination or nonvaccination groups.
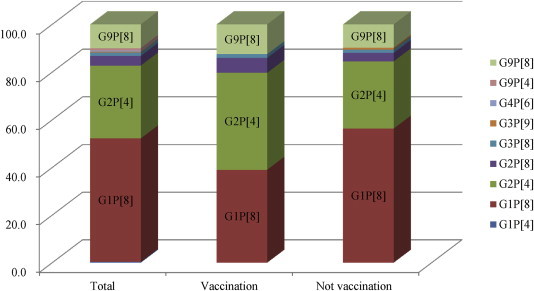

Table 1Frequency of rotavirus infection by vaccination status.
| Rotavirus | Vaccinated, n (%) | Not vaccinated, n (%) | p |
|---|---|---|---|
| Positive | 67 (17.4) | 144 (37) | 0.000 |
| Negative | 318 (82.6) | 245 (63) | |
| Total | 385 (100) | 389 (100) |
Table 2Distribution of rotavirus G and P genotypes in patients with acute gastroenteritic symptoms in Gwangju, South Korea in 2013.
Figure & Data
References
Citations
Citations to this article as recorded by 

- Rotavirus infection among hospitalized children under five years of age with acute watery diarrhea in Sri Lanka
Paba Palihawadana, Gagandeep Kang, Janakan Navaratnasingam, Geethani Galagoda, Janaki Abeynayake, Madhava Gunasekera, Shilanthi Seneviratne
Vaccine.2018; 36(51): 7846. CrossRef - Complete genome sequence analysis of rare G4P[6] rotavirus strains from human and pig reveals the evidence for interspecies transmission
Rungnapa Malasao, Pattara Khamrin, Kattareeya Kumthip, Hiroshi Ushijima, Niwat Maneekarn
Infection, Genetics and Evolution.2018; 65: 357. CrossRef - Post-marketing safety surveillance conducted in Korea (2008–2013) following the introduction of the rotavirus vaccine, RIX4414 (Rotarix™)
Son Moon Shin, Chun Soo Kim, Naveen Karkada, Aixue Liu, Girish Jayadeva, Htay Htay Han
Human Vaccines & Immunotherapeutics.2016; 12(10): 2590. CrossRef


 PubReader
PubReader Cite
Cite
