Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 2(3); 2011 > Article
-
Articles
Neutralizing Antibody Responses and Evolution of the Viral Envelope in the Course of HIV-1 Korean Clade B Infection - Bo Gyeong Shin, Mi-Ran Yun, Sung Soon Kim, Gab Jung Kim
-
Osong Public Health and Research Perspectives 2011;2(3):151-157.
DOI: https://doi.org/10.1016/j.phrp.2011.11.038
Published online: December 31, 2011
Division of AIDS, Korea National Institute of Health, Osong, Korea
- Corresponding author. E-mail: gabjkim@korea.kr
• Received: July 26, 2011 • Revised: September 30, 2011 • Accepted: October 27, 2011
Copyright ©2011, Korea Centers for Disease Control and Prevention
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License () which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Objectives
- HIV is able to continuously adapt to and evade the evolving neutralizing antibody responses of the host. We investigated the ability of HIV variants to evade neutralizing antibodies in order to understand the distinct characteristics of HIV-1 Korean clade B.
-
Methods
- Three drug-naive subjects were enrolled in this study who were infected with HIV-1 Korean clade B. Neutralizations were performed using autologous plasma and pseudovirion-based assays in order to analyze and compare changes in the env gene.
-
Results
- In the early phase of infection, neutralizing activities against autologous virus variants gradually increased, which was followed by a decline in the humoral immune response against the subsequent viral escape variants. The amino acids lengths and number of potential N-linked glycosylation sites (PNGS) in HIV-1 env gene was positively correlated with neutralized antibody responses during the early stages of infection.
-
Conclusion
- This study suggests that change within the env domains over the course of infection influences reactivities to neutralized antibodies and may also have an impact on host immune responses. This is the first longitudinal study of HIV-1 humoral immunity that took place over the entire course of HIV-1 Korean clade B infection.
- Within-host HIV-1 evolution is characterized by the diversification of the infecting virus population after the time of infection [1]. In addition to genetic drift and purifying selection, HIV-1 can evolve under positive selection from antibodies and cytotoxic T lymphocytes [2]. In this process, neutralizing antibodies are the principal component required for an effective immune response to HIV.
- HIV-1 envelope glycoprotein (Env) is the primary viral antigen that is targeted by neutralizing antibodies, so genetic variation in the env gene of HIV-1 is closely related to the neutralizing antibody response [3,4].
- Previous studies have reported that the development of Nab responses in the initial phase of HIV-1 infection [5,6], but there were few reports describing neutralization activities over the entire course of HIV evolution in longitudinally monitored HIV-positive subjects. Also, the relationship between neutralizing antibodies, HIV-1 genetic variation, and the functional mechanism by which neutralizing antibodies respond to HIV-1 is not completely understood. Therefore, in order to understand the evolution of the virus, a longitudinal study on viral variation and neutralizing antibody responses is need.
- Genetic diversity and divergence in HIV-1 Korean clade B are much lower than those reported in HIV strains in other countries [7]. These characteristics of HIV-1 Korean clade B may support important immunological advantages [8].
- The goal of this study is to investigate the relationship between Nab responses and the env gene, which is the target of neutralization responses in subjects with Nabs. We also examined sequential neutralization responses in autologous plasma obtained from patients infected with HIV-1 Korean clade B.
1. Introduction
- 2.1. Study subjects, cells, and plasma samples
- Blood samples taken from patients with a suspected HIV infection were referred to the Korea Center for Disease Control (KCDC) from public health centers, hospitals and local blood banks through local Institutes of Public Health and Environment (IPHE) for the final HIV confirmation test. Among these patients, three were diagnosed with preseroconversion status and could be monitored longitudinally. None of these subjects received antiretroviral therapy over the course of this study.
- 293T/17 and TZM-bl cells were obtained from the National Institute for Biological Standards and Control (catalog No. ARP5011) and the American Type Culture Collection (catalog No. 11268), respectively. An env-deficient HIV-1 backbone vector (pSG3 Env) was obtained through the NIH AIDS Research and Reference Reagent Program (catalog No. 11051).
- 2.2. Cloning of full-length HIV-1 env genes and pseudovirus production
- Genomic DNA was extracted from the peripheral blood mononuclear cells (PBMCs) using QIAamp DNA blood mini kits (QIAgen, Valencia, CA, USA). We amplified a 3.2-kb region of the HIV-1 env gene using nested polymerase chain reaction (PCR), as previously described [9].
- The purified PCR products were cloned into the pcDNA3.1/V5-His-Topo vector (Invitrogen Corp., Carlsbad, CA, USA). Pseudoviruses were produced by infecting 293 T cells with the env expression plasmid and pSGΔenv vector using the FuGENE 6 transfection kit (Invitrogen). Pseudovirus-containing culture supernatants were harvested 72 hours after transfection, filtered (0.45 ㎛), and stored at -80 ℃ until use in the neutralization assays.
- 2.3. Neutralization assay
- The activities of the neutralizing antibodies were measured as the reduction in β-galactosidase reporter gene expression after a single round of viral infection in TZM-bl cells, as previously described [1]. In brief, 100 TCID50 pseudoviruses and heat-inactivated plasma mixtures were incubated at 37℃ for 1 hour and then added to a preparation of TZM-bl cells (1 ×104 cells/mL) on 96-well plates. The cells were then harvested after incubation for 48 hours and counted using a β-galactosidase-staining V2600 staining kit (Takara Bio Inc., Tokyo, Japan). The 50% inhibitory concentration (IC50) of the neutralizing antibody was defined as the concentration of plasma dilution required to decrease the number of infected cells by 50% at 48 hours after infection with 100 TCID50, and the IC50 was calculated from the mean value of the repeated results. Two independent experiments were performed in duplicate.
- 2.4. Sequencing and phylogenetic analyses
- The sequencing reaction for the env region was performed using the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, CA, USA) in an automated ABI prism 3730 DNA sequencer (Perkin Elmer, CT, USA). The nucleotide and amino acid sequences of the env gene were aligned using the Lasergene software package (DNASTAR Inc., WI, USA). Phylogenetic analyses were performed using PAUP (Phylogenetic Analysis Using Parsimony, version 4.0; http://paup.csit.fsu.edu/). The number and position of potential N-linked glycosylation sites (PNGS) were analyzed using the N-GlycoSite program (www.hiv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html). Aligned amino acid sequences were analyzed using Jalview (www.jalview.org).
- 2.5. Statistical analysis
- Statistical analyses were performed using SAS version 9.1 (SAS Institute, Inc., Cary, NC, USA). Spearman’s rank correlation method was used to determine the significance of any association between the neutralizing antibody response and the HIV env sequence.
2. Materials and Methods
- 3.1. Characteristics of study subjects
- Three subjects (K1004, K1314, and K2160) with primary HIV infection (PHI) were selected for monitoring over the course of their infections, all of whom were male subjects. The average age of the selected group was 36.3 years. The average course of infection after PHI was 4.8 years, and eight time points were sampled over 85 months (7.1 years) for K1004, six time points over 63 months (5.3 years) for K1314, and five time points were sampled over 25 months (2.1 years) for K2160. The route of transmission for each subject was sexual contact with men who have sex with men (MSM). The mean values for the longitudinal samples were 449 cells/mm3 (range: 147-896 cells/mm3) for CD4+ T cells and 1,083 cells/mm3 (range: 599-1,954 cells/mm3) for CD8+ T cells (Table 1). Phylogenetic tree analysis of all HIV-1 env sequences showed that all isolates were HIV-1 Korean clade B (Figure 1).
- 3.2. Viral escape pattern of neutralization against autologous viruses over the course of evolution
- Pseudoviruses were produced from each individual in order to analyze neutralization reactivity. We determined the neutralization reactivity against autologous HIV-1 env pseudovirus at each sampled time point in order to analyze cross-reactivity at different stages of evolution.
- In the evaluations of the neutralizing activities to HIV-1 plasma at different stages, the pseudoviruses that were produced in the early phase showed low neutralizing reactivities to contemporaneous plasma compared with the later phase, which was significant except for K2160 (p < 0.005). In K1004 and K1314, gradual increases were observed after early phase, but these
- Epidemiological and immunological characteristics of Korean subjects infected with HIV-1 type B
- increased neutralization activities were reduced by 4 years post-HIV infection. K2160, who was monitored for relatively shorter time than the other subjects, showed a gradually increase in neutralization activities that was time dependent (Figure 2). Specifically, K1004, who produced by eight samples at each time point, showed the most complicated neutralization pattern which also showed reversion. There was an increase in the neutralization pattern in the early phase through the fourth time point, but this increase began to decrease through measurement at the seventh time point. In case of K1314, the results show an increased pattern from the first through the fifth time point and reduced neutralization by the last time point, similar to K1004. However, for K2160, who provided five samples at each time point, we were unable to determine the escape pattern in the later phase; only a steadily rising neutralization pattern was identified in the early phase.
- 3.3. Genetic variation of env gp160 over the course of evolution
- Genetic variations mainly existed in gp120 as result of insertions and deletions in this region. In K1004, most PNGS variations were observed in all variable loops over the course of infection, but some changes in the lengths of various amino acids could only be observed at V4 region (Table 2). In K1314, the lengths of the amino acids and PNGS were changed in all regions except V3. Specifically, the lengths of the amino acids over the course of infection showed a decrease in V1, V4, and V5, and the number of PNGS also decreased in V1 and
- V4. In K2160, variations in the lengths of amino acids were observed only at V1 and V5, but the number of PNGS changed in all regions except V3.
- In the analysis of the CD4- and CCR5-binding sites, most amino acids were conserved in those sites, but some changes were observed over the course of infection and the changes observed at each time point were different for each individual. Regarding changes in the CD4-binding site, D474 N was noted in subject K1004 and N279D, T283 N, and D474 N were noted in subject K1314. Regarding changes in CCR5-binding site, F317L and R440 K were noted in subject K1004, and changes in K440R were noted in subject K1314. However, no changes were observed in subject K2160 (data not shown).
- 3.4. Relationship between env sequence and neutralization reactivities
- The results of the analysis between variations in env genes and neutralization titers (IC50) showed that the lengths of the amino acids in regions V1-V5 had a significant but weakly positive correlation with the neutralization titers (r=0.279, p=0.026) in subject K1004. Also, the number of PNGS in same region was positively correlated with the neutralization titers (r=0.349, p=0.005). However, in the case of subject K1314, the lengths of the amino acids and the number of PNGS were weakly and positively correlated with the neutralization titers, but this finding was not significant. Also, correlation analysis of subject K2160 showed results that were similar to those of subject K1314 (Figure 3).
- Comparison of amino acids lengths and number of PNGS in HIV-1 env gp160 in Korean HIV-1 subjects
3. Results
Table 1.
Figure 1.

Phylogenetic relationships between HIV-1 env (gp120) sequences of Korean subjects infected by HIV-1 Korean clade B and env sequences of 17 different type B reference strains from the HIV database in Los Alamos National Laboratory (http://www.hiv.lanl.gov). This phylogenetic tree was constructed using the neighbor-joining method used by the PAUP* 4.10b program. The numbers at the nodes represent the bootstrap values calculated from 1000 bootstrap resamplings. The three infected Korean patients are labeled from according to the sampling time.

Figure 2.

Autologous cross-reactivity of HIV-1 pseudoviruses against neutralizing antibodies in Korean HIV-1 longitudinal subjects over the course of HIV-1 infection. PV, pseudovirus.

Table 2.
- This study was designed to investigate the mechanism used by HIV-1 Korean clade B to escape humoral immune responses.
- In order to respond to the HIV virus, broadly reactive neutralization responses against the virus must be developed in order to design an effective HIV-1 vaccine. Therefore, it is important to investigate the genetic characteristics of predominant HIV strains in each country and understand the subsequent immune mechanisms involved in the neutralizing antibody responses.
- As previously reported, the expansion of genetic variations in HIV-1 infected Korean subjects is much lower than that reported in other countries [7]. Also, the neutralizing antibody responses to HIV-1 Korean clade B show broad reactivity to HIV-1 subtype B [8]. In order to develop effective, region-specific vaccines, understanding the limited genetic characteristics of Korean clade B might be advantageous to understanding how HIV elicits humoral immune responses.
- In this study, all subjects developed neutralizing antibody responses against their sequential autologous plasma. Within most time points, titers against an earlier virus were greater than against contemporaneous or later viruses, indicating changes in immune pressure and viral escape.
- Similar to other observations [1,4,10], two of the three subjects showed a decline in autologous neutralization during chronic infection as a result of both viral
- escape and the limited ability of the immune response against viral escape variants. Thus, the loss of neutralization by sequential viruses over time suggests that some viruses could evolve to escape the effects of a potent vaccine, and vaccines must target the envelope regions that are less likely to evolve and escape recognition by vaccine-induced antibodies [4].
- Other studies have shown that HIV-1 sequences that have been isolated during chronic infection have significantly longer V1-V2 loops and a significantly higher number of PNGS sites than sequences that are isolated during the early phases of infection [11,12]. These studies suggest that changes within the V1-V2 envelope domains over the course of an infection influences sensitivity to autologous neutralizing antibodies and may also have an impact on host interactions. However, we observed a decrease in the lengths of the variable loops and in the number of PNGS in gp120 that were irrespective of the detected autologous neutralizing activities in plasma. Also, PNGS analysis demonstrated only a small amount of variation, but in a subjectspecific way, suggesting that these variations are not confined to the variable regions of gp120 and can also be observed in constant regions.
- In our study, Nab responses did not correlate with env variation during chronic infection. The absence of an absolutely temporal relationship between the dynamics of the length and level of glycosylation of env indicate that the influence of the selection processes on Env characteristics may be caused by various factors. Especially, the escape pattern from neutralizing antibody responses may occur through a combination of point mutations (i.e., substitution of amino acids), changes in glycosylation patterns, and insertions and deletions in the viral envelope [13]. This suggests that the Nab response is not a dominant, selective force on env evolution.
- Additionally, these characteristics of HIV is based on many complicated mechanisms and factors, such as the high diversity of HIV and the structural features that determine T cell accessibility and glycosylation that might affect the HIV-neutralization epitope; therefore, HIV evolution should be considered when studying HIV neutralization. One study on HIV subtype C [12] showed that the escape pattern of HIV might be influenced by immune responses to different regions and that the HIV neutralization process requires multiple pathways.
- In conclusion, this is the first report on neutralizing humoral immune responses against HIV-1 Korean clade B and viral escape that take into account the molecular changes in the viral envelope that take place over the entire course of infection. HIV-1 Korean clade B viruses can evolve over time to become resistant or sensitive to the effects of antibodies, so the changes in Env should be carefully considered when attempting to design a potent vaccine against HIV-1 Korean clade B. This information may be useful for developing an AIDS vaccine and therapies against HIV-1 Korean clade B.
4. Discussion
Figure 3.
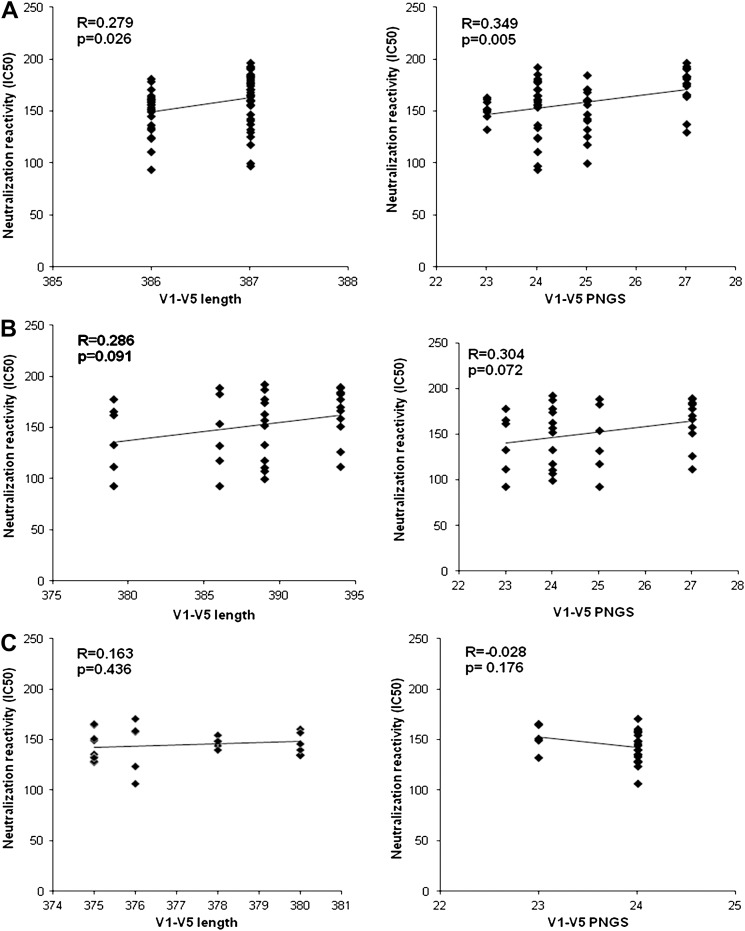
Correlation between amino acid length and PNGS number in the V1–V5 regions of HIV-1-infected Korean subjects and the total reactivity against neutralizing antibodies. (A) K1004, (B) K1314, (C) K2160.

-
Acknowledgements
- This research was supported by an intramural grant from the Korea National Institute of Health (2007-N51002-00).
- 1. Wei X Decker JM Wang S et al.. Antibody neutralization and escape by HIV-1. Nature 20 3 2003;422(6929). 307−12. PMID: 12646921.ArticlePubMed
- 2. Rambaut A Posada D Crandall KA Holmes EC . The causes and consequences of HIV evolution. Nat Rev Genet 1;2004;5:52−61. PMID: 14708016.ArticlePubMed
- 3. Nyambi P Burda S Urbanski M et al.. Neutralization patterns and evolution of sequential HIV type 1 envelope sequences in HIV type 1 subtype B-infected drug-naïve individuals. AIDS Res Hum Retroviruses 12;2008;24(12). 1507−19. PMID: 19018670.ArticlePubMedPMC
- 4. Bunnik EM Pisas L van Nuenen AC Schuitemaker H . Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J Virol 8;2008;82(16). 7932−41. PMID: 18524815.ArticlePubMedPMC
- 5. Frost SD Wrin T Smith DM et al.. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci USA 20 12 2005;102(51). 18514−9. PMID: 16339909.ArticlePubMedPMC
- 6. Richman DD Wrin T Little SJ Petropoulos CJ . Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA 4;2003;100(7). 4144−9. PMID: 12644702.ArticlePubMedPMC
- 7. Kim GJ Nam JG Shin BG et al.. National survey of prevalent HIV strains: limited genetic variation of Korean HIV-1 clade B within the population of Korean men who have sex with men. J Acquir Immune Defic Syndr 6;2008;48(2). 127−32. PMID: 18317230.ArticlePubMed
- 8. Shin BG Kim SS Kim GJ . Broad neutralizing antibody response and genetic variation in HIV-1 env genes in Koreans with primary HIV-1 infections. Arch Virol 3;2011;153(3). 465−72. PMID: 21184245.Article
- 9. Li M Gao F Mascola JR et al.. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 8;2005;79(16). 10108−25. PMID: 16051804.ArticlePubMedPMC
- 10. Rong R Li B Lynch RM et al.. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathogens 9;2009;5(9). e1000594PMID: 19763269.ArticlePubMedPMC
- 11. Chohan B Lang D Sagar M et al.. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol 5;2005;79(10). 6528−31. PMID: 15858037.ArticlePubMedPMC
- 12. Sagar M Wu X Lee S Overbaugh J . Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J Virol 10;2006;80(19). 9586−98. PMID: 16973562.ArticlePubMedPMC
- 13. Haldar B Burda S Williams C et al.. Longitudinal study of primary HIV-1 isolates in drug-naive individuals reveals the emergence of variants sensitive to anti-HIV-1 monoclonal antibodies. PLoS One 23 2 2011;6(2). e17253PMID: 21383841.ArticlePubMedPMC
Figure & Data
References
Citations
Citations to this article as recorded by 

- Blockage of CD59 Function Restores Activities of Neutralizing and Nonneutralizing Antibodies in Triggering Antibody-Dependent Complement-Mediated Lysis of HIV-1 Virions and Provirus-Activated Latently Infected Cells
Kai Yang, Jie Lan, Nicole Shepherd, Ningjie Hu, Yanyan Xing, Daniel Byrd, Tohti Amet, Corlin Jewell, Samir Gupta, Carole Kounga, Jimin Gao, Qigui Yu, G. Silvestri
Journal of Virology.2015; 89(18): 9393. CrossRef


 PubReader
PubReader Cite
Cite
