Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 3(1); 2012 > Article
-
Articles
Phylogenetic Analysis of the Rotavirus Genotypes Originated from Children < 5 Years of Age in 16 Cities in South Korea, between 2000 and 2004 - Ho-Kyung Oha,b, Seung-Hwa Honga, Byung-Yoon Ahnb, Hye-Kyoung Minc
-
Osong Public Health and Research Perspectives 2011;3(1):36-42.
DOI: https://doi.org/10.1016/j.phrp.2012.01.006
Published online: December 31, 2011
aNational Center for Lot Release, National Institute of Food & Drug Safety Evaluation, Korea Food & Drug Administration, Osong, Korea.
bSchool of Life Sciences and Biotechnology, Korea University, Seoul, Korea.
cDepartment of Biopharmaceuticals & Herbal Medicine Evaluation, Korea Food & Drug Administration, Osong, Korea.
- Corresponding author. E-mail: bsmin@korea.kr
• Received: December 8, 2011 • Revised: January 15, 2012 • Accepted: January 20, 2012
Copyright ©2012, Korea Centers for Disease Control and Prevention
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License () which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Objectives
- The purpose of this study was to examine the diversity of the G and P types of human rotavirus strains isolated in South Korea during 2000 to 2004.
-
Methods
- We selected 38 Group A rotavirus isolates among 652 fecal samples, which were collected from infants and children < 5 years of age with acute gastroenteritis or diarrhea admitted in 8 hospitals representative of five provinces of South Korea between 2000 and 2004. Rotavirus P- and G-genotypes were determined by nucleotide sequencing and phylogenetic analysis was performed.
-
Results
- One G1P[4] consisted G1-Id-P[4]-V; one G1P[6] consisted G1-Id-P[6]-Ia; nine G1P[8] consisted G1-Ib-P[8]-Ia (n=3), G1-Ic-P[8]-Ia (n=1), and G1-Id-P[8]-Ia (n=5); 13 G2P[4] consisted G2-V-P[4]-V; two G3P[4] consisted G3-IIId-P[4]-V; five G3P[8] consisted G3-IIId-P[8]-Ia; four G4P[6] consisted G4-Ie-P[6]-Ia; two G4P[8] consisted G4-Ie-P[8]-II; one G9P[6] consisted G9-III-P[6]-Ia.
-
Conclusions
- A considerable amount of rotavirus genotypic diversity was detected in South Korea from 2000 to 2004. These findings are important to develop the effective vaccines and to undertake epidemiologic studies.
- Group A rotavirus, the most common etiologic agent of severe diarrhea in children, causes about 600,000 deaths per year [1]. Rotavirus, which is a genus belonging to the Reoviridae family, has a genome of 11 segments of double-stranded RNA surrounded by a triple-layered capsid consisting of a core, inner capsid, and outer capsid. The outer capsid is composed of two structural proteins, VP4 and VP7, which define virus G(VP4) or P(VP7) serotype specificity [2]. Although at least 15 G genotypes and 26 P genotypes are known [3-8], the most prevalent P-G combinations in humans are G1P[8], G2P[4], G3P[8], G4P[8] and G9P[8]. In Korea, rotavirus is still the most common viral agent of acute diarrhea in young children. Although G1P[8] was the most prevalent strain until 1997 regardless of geographic area or season [6,9], the predominant G type strain has shifted to other genotypes including G4, G2 or G9 [10-13]. In the present study, we examined the diversity of the G and P types of human rotavirus strains isolated in South Korea during 2000 – 2004 periods. As a result, we confirmed that total nine P-G genotypic isolates were identified.
1. Introduction
- 2.1. Sample collection
- A total of 38 rotavirus isolates were selected among 652 fecal samples, which were collected from infants and children < 5 years of age with acute gastroenteritis or diarrhea admitted in eight hospitals representative for five provinces of South Korea between 2000 and 2004. Human rotaviruses were detected in 354 of 652 (54.3%) fecal samples by enzyme-linked immunosorbent assay (ELISA). G and P genotypes were detected by multiplex polymerase chain reaction (PCR) in 316 (89.3%) and 327 (92.4%) of these sample, respectively. The location of these areas was plotted on the map of South Korea is shown Figure 1.
- 2.2. Nucleotide sequencing
- Human rotavirus (HRV) double-stranded RNA (dsRNA) was extracted using a QIAamp Viral RNA kit (Qiagen GmbH, Hilden, Germany) in accordance with the manufacturer’s instructions. The dsRNA samples
- were subjected to seminested multiplex Reverse Transcriptase- Polymerase Chain Reaction (RT-PCR) using conserved and type specific primers (VP7-G1, G2, G3, G4, and G9, and VP4-P[4], P[6], and P[8])[14-18]. The PCR amplicons were purified using a commercial spin column method (Qiagen GmbH, Hilden, Germany) and sequenced automatically using the ABI PRISM 3100 automated DNA sequencer (Applied Biosystems, Inc, Foster city, California, USA).
- 2.3. Phylogenetic analysis of nucleotide sequences
- The VP4 and VP7 sequences obtained were aligned and compared with others VP4 and VP7 sequences of rotaviruses available in the Genebank database (http://www.ncbi.nlm.nih.gov/genbank/). Phylogenetic trees of alignment were constructed using the neighbor-joining method by bootstrapping with 1000 replicates and phylogenetic distances were measured by the Tajima-Nei model [19] implements in the Molecular Evolutionary Genetics Analysis (MEGA) (http://www.megasoftware.net/) analytical package (version 5.05, Institute of Molecular Evolutionary Genetics and Department of Biology, Pennsylvania State University, University Park, Pennsylvania state, USA).
- 2.4. Nucleotide sequence accession numbers
- The VP4 nucleotide sequences of the human rotavirus in this study were assigned accession numbers EF015885, EF077317-EF077322, EF077324-EF077342, EF077344, EF077345, and EF0773347-EF077356. The VP7 nucleotide sequences were deposited in NCBI Gen-Bank under accession numbers HQ425255-HQ425292. The referenced sequences in the GenBank database are shown in Table 1.
2. Methods
Figure 1.

Location of the five provinces used in this study. Numbers in parentheses indicate the number of case.

- Most of the fecal samples were collected from Gyunggi province (306/652), followed by Gyeongsang province (130/652), Chungcheong province (88/652), Gangwon province (85/652), Jeolla province (43/652). These are shown in Figure 1. Among 652 stool samples, 354 (54.3%) children below 5 years of age were reactive in ELISA. G and P genotypes in parallel were detected by multiplex PCR in 314(88.7%) of these samples.
- During the 2000–2004 period, nine P-G combinations (N=314) are prevalent in South Korea. Overall, G2P[4](53.5%) was the most dominant, followed by G1P[8](16.6%), G1P[6](13.7%), G4P[6](7.3%), G4P[8](5.7%), G3P[8](1.3%), G3P[4](1.0%), G1P[4] (0.6%) and G9P[6] (0.3%), which are shown Table 2. G2P[4] [Gyunggi (47.0%), Gangwon (58.1%), Chungcheong (46.9%), Jeolla (36.8%), and Gyeongsang (70.4%)] was the dominant combination of genotypes. However, subdominant combinations were different in five provinces. G1P[6] and
- Reference sequence of this study
- G1P[8] prevailed in Gyunggi province (20.1%, 17.2%), G1P[8] prevailed in Gangwon province (25.8%), G1P[8], G4P[6] and G4P[8] prevailed in Chungcheong province (18.4%, 14.3% and 14.3%), and G1P[6] prevailed in Gyeongsang province (13.6%). Also, G1P[8] and G3P[8] prevailed in Jeolla province.
- To examine the VP7 and VP4 nucleotide sequences for 38 isolates among 314 identified isolates, phylogenetic trees for G type (G1, G2, G3, G4, and G9) and P type [P(4), P(6), P(8)) were constructed by applying the neighbor-joining method. Sequences of VP7 and VP4 were determined from 38 representative rotaviruses, comprising the different genotypes and intra genotypic lineages detected by partial sequencing. Sequences of the representative isolates were submitted to GenBank (Table 3) and included in the phylogenetic analysis (Figures 2 and 3). In this study, the 11 G1 rotavirus isolates showed that they are a part of the lineage I and are clustered into five
- Distribution of group A rotavirus P-G combination strains among infants and children below 5 years of age with diarrhea in five provinces of South Korea between 2000 and 2004
- minor lineages(Ia-Ie) in the phylogenetic analysis. The G1 rotaviruses segregated into seven major lineages (I–VII ) as reported by Arista and colleagues [20]. Most of these isolates are clustered in sub-lineage Id (n=7), followed by sublineage Ib (n=3) and sublineage Ic (n=1). Sublineage Id isolates showed 97.8%~99.2% nucleotide sequence similarity to strain GER15-08, and sublineage Ib isolates showed 99.3%~99.6%nucleotide similarity with strains VN-281.KMR267 isolates in sublineage Ic showed 99.5% nucleotide similarity to Py9856. Among eleven G1 rotavirus isolates, the three G1-Ib isolate was associated with P[8]-IIIa. One G1-Ic was associated with P[8]-IIIa and seven G1-Id isolates were associated with P[4]-V, P[6]-Ia, and P[8]-IIIa.
- The G2 rotaviruses segregated into five major lineages (I–V) [21]. The thirteen G2 rotavirus isolates clustered under lineage V. They showed 99.2~99.5% nucleotide similarity to strain GER84-08 from Germany. All G2-V
- The G and P genotypes of the 38 representative rotavirus strains of this study are given. The VP7 and VP4 sequences were submitted to GenBank and the accordant accession numbers are provided in brackets. atient age, gender, city of sample collection, and the year of sample collection are indicated
- isolates were associated with P[4]-V. Meanwhile, the seven G3 strains are a part of the sublineage IIId and showed 99.3%~99.5% sequence similarity to strain GER198-08. The two G3-IIId were associated with P[4]-V, and five G3-IIId were associated with P[8]-IIIa. The 16 P[4]-V including G1-Id, G2-V, and G3-IIId shared more than 97.6% nucleotide similarity, and the six G4 rotavirus strains clustered in sub-lineage Ie compared to strains KUMS00-74 has 98.7-99.4% nucleotide similarity. The four G4-Ie were associated with P[6]-Ia and two G4-Ie were associated with P[8]-II. Finally, the one G9 isolate, KMR720 showed high sequence similarity to all lineage III strains (more than 98.3%) and associated with P[6]-Ia. While the five P[6]-Ia excluding G9 isolate exhibited 98.4%~98.7% nucleotide sequence similarity to TE56, G9 isolate, KMR720 exhibited 97.1% nucleotide sequence similarity to GR846/86. While P[8]-IIIa exhibited 97.6%~ 99.4% nucleotide similarity to strains CMH032/05, P[8]-II exhibited 99.4% nucleotide sequence similarity to strain Kagawa/88-104. As a result, we confirm that a total of nine P-G genotypic isolates were identified (11 P-G subgenotypes).
3. Results
Table 1.
Table 2.
Table 3.
- Rotaviruses have been described as a major cause of severe diarrhea among infants and young children in
- South Korea. Epidemiologic studies worldwide have revealed that five P-G combinations, G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8], have been linked to most of the cases of rotavirus diarrhea among infants and young children worldwide. In this study, we confirmed both uncommon P-G combinations [G1P(4), G1P(6), G3P(4), G4P(6), and G9P(6)] as well as most common P-G combinations [G1P(8), G2P(4), G3P(8), and G4P(8))] There are two oral live vaccines available in Korea; Rotarix (GlaxoSmithKline, Rixensart, Belgium) is a monovalent vaccine that consists of the attenuated G1P[8] human rotavirus strain RIX4414 and has been in use in Korea since 2008. Rotateq (MERCK & CO.,INC, Pennsylvania, USA) is a pentavalent vaccine which consists of five attenuated human-bovine reassortant viruses [G1 to G4 and P(8)]; G1: human W179-bovine WC3 reassortant, G2: human SC2-bovine WC3 reassortant, G3: W178-bovine WC3 reassortant, G4: human BrB-bovine WC3 reassortant, P1A[8]: human W179-bovine WC3 reassortant. It has been available in Korea since 2007. Although these vaccines are effective on most common P-G combinations, G2P[4], G1P[8], G3P[8], G4P[8], and G9P[8], those may fail to work on the other genotypes. All things considered, development of multivalent rotavirus vaccine including new ones must be required. Therefore, this study will provide useful information for the development of effective rotavirus vaccines in the future. Also, the international relationship has been limited by the geographic location in the past, but now it has expanded all over the world, and this expansion is thought to be the cause of the change in strains.
4. Discussion
Figure 2.
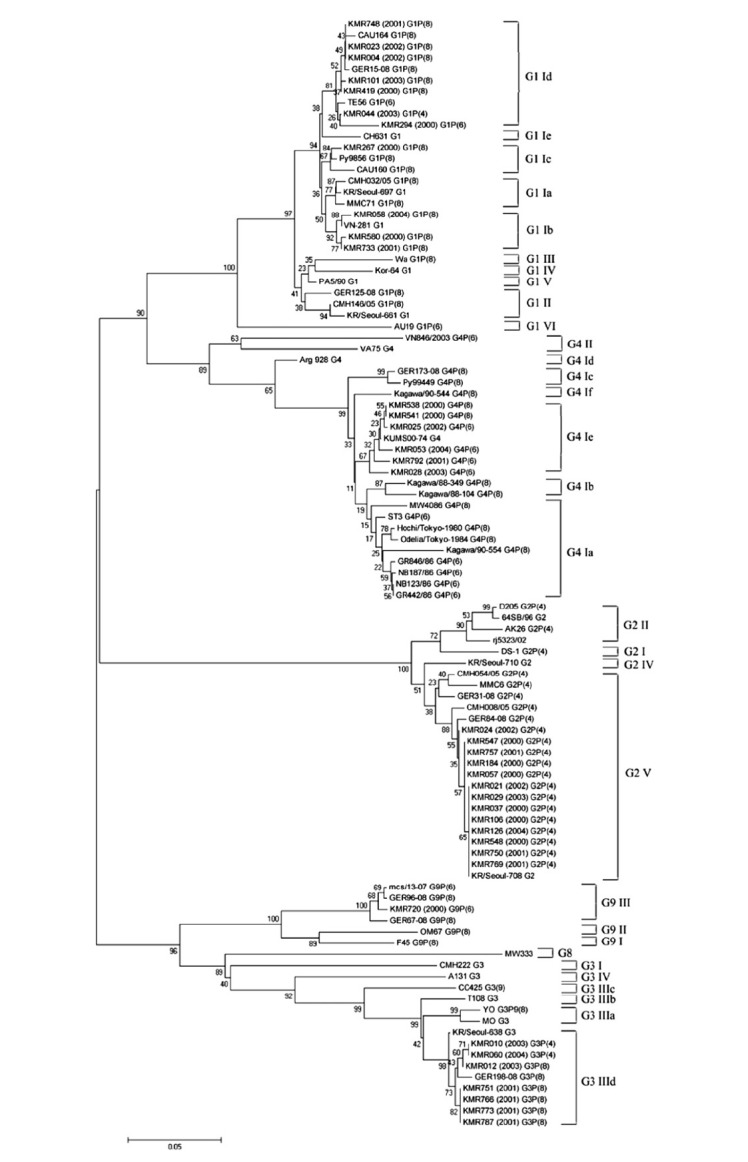
Phylogenetic analysis of VP7 gene nucleotide sequences of Group A rotavirus strains from Korea between 2000 and 2004. Phylogenetic trees of alignment were constructed using the neighbor-joining method by bootstrapping with 1000 replicates, and phylogenetic distances were measured by Tajima-Nei model. Only values > 50% are given. Numbers at nodes indicate the level of bootstrap support (%). Bar represents 0.05 substitutions per nucleotide position.

Figure 3.
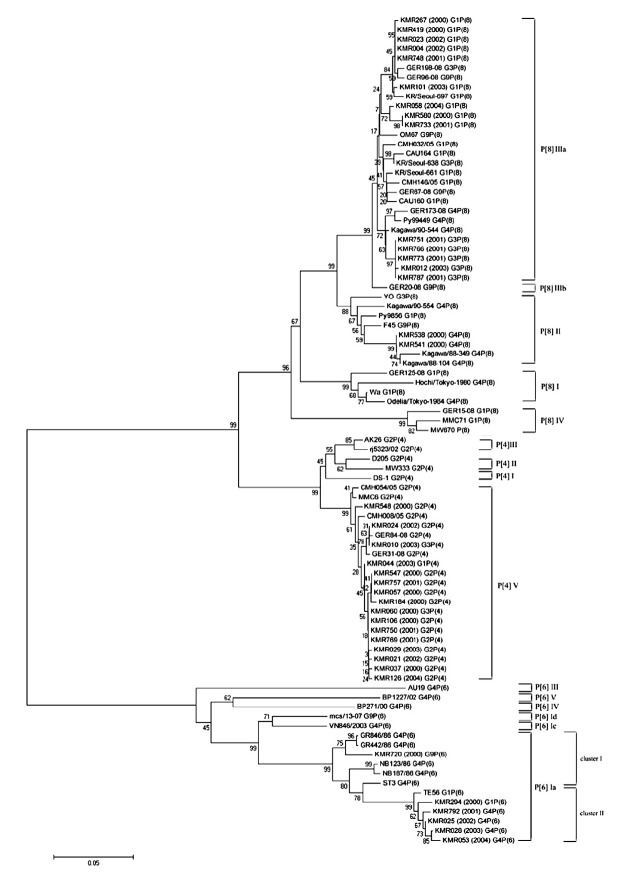
Phylogenetic analysis of VP4 gene nucleotide sequences of Group A rotavirus strains from Korea between 2000 and 2004. Phylogenetic trees of alignment were constructed using the neighbor-joining method by bootstrapping with 1000 replicates, and phylogenetic distances were measured by Tajima-Nei model. Only values > 50% are given. Numbers at nodes indicate the level of bootstrap support (%). Bar represents 0.05 substitutions per nucleotide position.

- Consequently, the P-G combination genotypes of this study would serve as useful information for the development of effective rotavirus vaccines. Such P-G combinational phylogenetic study will also be necessary for international epidemiologic investigation of human rotaviruses providing novel insights into the interspecies transmission processes of rotaviruses. To strengthen our opinion, we highlight the need for continued monitoring of circulating rotavirus strains for effective prevention and vaccine development strategies.
5. Conclusion
-
Acknowledgements
- This study was supported in part by a grant from the KFDA Research and Development Program on Strengthening the Safety of Biological Products.
- 1. Parashar UD Gibson CJ Bresse JS Glass RI. . Rotavirus and severe childhood diarrhea. Emerg Infect Dis 2;2006;12(2). 304−6. PMID: 16494759.ArticlePubMedPMC
- 2. Zhou YJ Burns JW Morita Y et al.. Localization of rotavirus VP4 neutralization epitopes involved in antibody-induced conformational changes of virus structure. J Virol 6;1994;68(6). 3955−64. PMID: 7514681.ArticlePubMedPMC
- 3. Rahman M Banik S Faruque ASG et al.. Detection and characterization of human group C rotaviruses in Bangladesh. J Clin Microbiol 9;2005;43(9). 4460−5. PMID: 16145092.ArticlePubMedPMC
- 4. Santos N Hoshino Y. . Global distribution of rotavirus serotypes/ genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol 1;2005;15(1). 29−56. PMID: 15484186.ArticlePubMed
- 5. Samajdar S Varghese V Barman P et al.. Changing pattern of human group A rotaviruses: emergence of G12 as an important pathogen among children in eastern India. J Clin Virol 7;2006;36(3). 183−8. PMID: 16679056.ArticlePubMed
- 6. Moon SS Song Green YS Song JW et al.. Genetic distribution of group A human rotavirus types isolated in Gyunggi province of Korea, 1999-2002. J Clin Virol 1;2007;38(1). 57−63. PMID: 17118702.ArticlePubMed
- 7. Varshney B Jagannath MR Vethanayagam RR et al.. Prevalence of, and antigenic variation in, serotype G10 rotaviruses and detection of serotype G3 strains in diarrheic calves: implications for the origin of G10P11 or P11 type reassortant asymptomatic strains in newborn children in India. Arch Virol 1;2002;147(1). 143−65. PMID: 11855628.ArticlePubMed
- 8. Martella V Banyai K Ciarlet M et al.. Relationships among porcine and human P[6] rotaviruses: evidence that the different human P[6] lineages have originated from multiple interspecies transmission events. Virology 1;2006;344(2). 509−19. PMID: 16194556.ArticlePubMed
- 9. Le VP Chung YC Kim KJ et al.. Genetic variation of prevalent G1P[8] human rotaviruses in South Korea. J Med Virol 5;2010;82(5). 886−96. PMID: 20336735.ArticlePubMed
- 10. Min BS Noh YJ Shin JH et al.. Surveillance study (2000 to 2001) of G- and P-type human rotaviruses circulating in South Korea. J Clin Microbiol 9;2004;42(9). 4297−310. PMID: 15365026.ArticlePubMedPMC
- 11. Kang JO Kilgore P Kim JS et al.. Molecular epidemiological profile of rotavirus in South Korea, July 2002 through June 2003: emergence of G4P[6] and G9P[8] strains. J Infect Dis 9;2005;192(Suppl. 1). S57−63. PMID: 16088806.ArticlePubMed
- 12. Kim JS Kang JO Cho SC et al.. Epidemiological profile of rotavirus infection in the Republic of Korea: results from prospective surveillance in the Jeongeub District, 1 July 2002 through 30 June 2004. J Infect Dis 9;2005;192(Suppl. 1). S49−56. PMID: 16088805.ArticlePubMed
- 13. Song MO Kim KJ Chung SI et al.. Distribution of human group a rotavirus VP7 and VP4 types circulating in Seoul, Korea between 1998 and 2000. J Med Virol 6;2003;70(2). 324−8. PMID: 12696125.ArticlePubMed
- 14. Das BK Gentsch JR Cicirello HG et al.. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 8;1994;32(7). 1820−2. PMID: 7929782.ArticlePubMedPMC
- 15. Gouvea V Santos N Timenetsky MC. . VP4 typing of bovine and porcine group A rotaviruses by PCR. J Clin Microbiol. 5;1994;32(5). 1333−7. PMID: 8051262.ArticlePubMedPMC
- 16. Wu H Taniguchi K Wakasugi F et al.. Survey on the distribution of the gene 4 alleles of human rotaviruses by polymerase chain reaction. Epidemiol Infect 1;1994;112(3). 615−22. PMID: 8005227.ArticlePubMedPMC
- 17. Gentsch JR Galss RI Woods P et al.. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 6;1992;30(6). 1365−73. PMID: 1320625.ArticlePubMedPMC
- 18. Taniguch K Wakasugi F Pongsuwanna Y et al.. Identification of human and bovine rotavirus serotypes by polymerase chain reaction. Epidemiol Infect 10;1992;109(2). 303−12. PMID: 1327857.ArticlePubMedPMC
- 19. Tajima F Nei M. . Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol 4;1984;1(3). 269−85. PMID: 6599968.PubMed
- 20. Arista S Giammanco GM De Grazia S et al.. Heterogeneity and temporal dynamics of evolution of G1 human rotaviruses in a settled population. J Virol 11;2006;80(21). 10724−33. PMID: 16928744.ArticlePubMedPMC
- 21. Khamrin P Maneekarn N Peerakome S et al.. Novel porcine rotavirus of genotype P[27] shares new phylogenetic lineage with G2 porcine rotavirus strain. Virology 5;2007;361(2). 243−52. PMID: 17215015.ArticlePubMed
Figure & Data
References
Citations
Citations to this article as recorded by 

- Long-term monitoring of G1P[8] rotaviruses circulating without vaccine pressure in Nizhny Novgorod, Russia, 1984-2019
N. A. Novikova, T. A. Sashina, N. V. Epifanova, A. U. Kashnikov, O. V. Morozova
Archives of Virology.2020; 165(4): 865. CrossRef - Prevalence, risk factors, and clinical characteristics of rotavirus and adenovirus among Lebanese hospitalized children with acute gastroenteritis
Rasha Zaraket, Ali Salami, Marwan Bahmad, Ali El Roz, Batoul Khalaf, Ghassan Ghssein, Hisham F. Bahmad
Heliyon.2020; 6(6): e04248. CrossRef - Prevalence and Genotypic Distribution of Rotavirus in Thailand: A Multicenter Study
Pimmada Sakpaisal, Sasikorn Silapong, Amara Yowang, Gaysorn Boonyasakyothin, Boonyaorn Yuttayong, Umaporn Suksawad, Siriporn Sornsakrin, Paphavee Lertsethtakarn, Ladaporn Bodhidatta, John M. Crawford, Carl J. Mason
The American Journal of Tropical Medicine and Hygi.2019; 100(5): 1258. CrossRef - Monitoring Shedding of Five Genotypes of RotaTeq Vaccine Viruses by Genotype-Specific Real-Time Reverse Transcription-PCR Assays
Yuki Higashimoto, Masaru Ihira, Yu Miyazaki, Ayumi Kuboshiki, Sayaka Yoshinaga, Hiroyuki Hiramatsu, Ryota Suzuki, Masafumi Miyata, Hiroki Miura, Satoshi Komoto, Jun Yukitake, Koki Taniguchi, Yoshiki Kawamura, Tetsushi Yoshikawa, Yi-Wei Tang
Journal of Clinical Microbiology.2018;[Epub] CrossRef - Molecular analysis of group A rotaviruses detected in hospitalized children from Rawalpindi, Pakistan during 2014
Massab Umair, Bilal Haider Abbasi, Nadia Nisar, Muhammad Masroor Alam, Salmaan Sharif, Shahzad Shaukat, Muhammad Suleman Rana, Adnan Khurshid, Ghulam Mujtaba, Uzma Bashir Aamir, Syed Sohail Zahoor Zaidi
Infection, Genetics and Evolution.2017; 53: 160. CrossRef - Analysis of rotavirus genotypes in Korea during 2013: An increase in the G2P[4] genotype after the introduction of rotavirus vaccines
Jae-Seok Kim, Hyun Soo Kim, Jungwon Hyun, Han-Sung Kim, Wonkeun Song, Kyu Man Lee, Seon-Hee Shin
Vaccine.2014; 32(48): 6396. CrossRef


 PubReader
PubReader Cite
Cite
