Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 14(4); 2023 > Article
-
Original Article
Increased viral load in patients infected with severe acute respiratory syndrome coronavirus 2 Omicron variant in the Republic of Korea -
Jeong-Min Kim1
 , Dongju Kim1
, Dongju Kim1 , Nam-Joo Lee1
, Nam-Joo Lee1 , Sang Hee Woo1
, Sang Hee Woo1 , Jaehee Lee1
, Jaehee Lee1 , Hyeokjin Lee1
, Hyeokjin Lee1 , Ae Kyung Park1
, Ae Kyung Park1 , Jeong-Ah Kim1
, Jeong-Ah Kim1 , Chae Young Lee1
, Chae Young Lee1 , , Il-Hwan Kim1
, , Il-Hwan Kim1 , Cheon Kwon Yoo2
, Cheon Kwon Yoo2 , Eun-Jin Kim1
, Eun-Jin Kim1
-
Osong Public Health and Research Perspectives 2023;14(4):272-278.
DOI: https://doi.org/10.24171/j.phrp.2023.0024
Published online: July 27, 2023
1Division of Emerging Infectious Diseases, Bureau of Infectious Disease Diagnosis Control, Korea Disease Control and Prevention Agency, Cheongju, Republic of Korea
2Bureau of Infectious Disease Diagnosis Control, Korea Disease Control and Prevention Agency, Cheongju, Republic of Korea
- Corresponding author: Eun-Jin Kim Division of Emerging Infectious Diseases, Bureau of Infectious Disease Diagnosis Control, Korea Disease Control and Prevention Agency, 187 Osongsaengmyeong 2-ro, Osong-eup, Heungdeok-gu, Cheongju 28159, Republic of Korea E-mail: ekim@korea.kr
© 2023 Korea Disease Control and Prevention Agency.
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
- 1,345 Views
- 108 Download
Abstract
-
Objectives
- Coronavirus disease 2019 (COVID-19) has been declared a global pandemic owing to the rapid spread of the causative agent, severe acute respiratory syndrome coronavirus 2. Its Delta and Omicron variants are more transmissible and pathogenic than other variants. Some debates have emerged on the mechanism of variants of concern. In the COVID-19 wave that began in December 2021, the Omicron variant, first reported in South Africa, became identifiable in most cases globally. The aim of this study was to provide data to inform effective responses to the transmission of the Omicron variant.
-
Methods
- The Delta variant and the spike protein D614G mutant were compared with the Omicron variant. Viral loads from 5 days after symptom onset were compared using epidemiological data collected at the time of diagnosis.
-
Results
- The Omicron variant exhibited a higher viral load than other variants, resulting in greater transmissibility within 5 days of symptom onset.
-
Conclusion
- Future research should focus on vaccine efficacy against the Omicron variant and compare trends in disease severity associated with its high viral load.
- The World Health Organization (WHO) tracks variants of concern, variants of interest, and variants under monitoring of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). These variants are classified according to their impact on public health. The Delta and Omicron variants have been classified as variants of concern, and each has distinct characteristics in terms of transmission, pathogenicity, and immune evasion mechanisms [1]. These differences are related primarily to amino acid substitutions in the receptor-binding domain, which facilitate the binding of the viral spike (S) protein to host cells [2]. The Omicron variant was first reported in South Africa on November 24, 2021, and was classified as a variant of concern by the WHO Technical Advisory Group on Virus Evolution [3,4]. The Omicron variant surged to global predominance, and the first confirmed case of infection in the Republic of Korea was reported on December 1, 2021. Currently, most coronavirus disease 2019 (COVID-19) cases globally are caused by sublineages of Omicron [5,6].
- The Omicron variant is classified as including more than 113 sublineages, according to the Phylogenetic Assignment of Named Global Outbreak Lineages (PANGOLIN) website. Thirty-two mutations have been identified in the S protein, including 15 amino acid substitutions in the receptor-binding domain, leading to changes in its viral characteristics, such as increased transmissibility and immune evasion [7,8]. The mutational signature of the Omicron variant has resulted in high transmission levels, contributing significantly to global outbreaks [9,10]. Therefore, to prevent and control additional widespread public health crises, it is important to understand the properties of the Omicron variant, including its transmissibility, virulence, immune response, and disease severity.
- The BA.2 sublineage of the Omicron variant spread rapidly in February 2022 [11]. BA.2 has a growth advantage over BA.1; it is also more transmissible than BA.1 and was the most widely reported Omicron sublineage internationally during the first half of 2022. Furthermore, a Japanese study has shown that BA.2 is associated with a severe disease in hamsters [12]. The study has also demonstrated strong protection against BA.2 reinfection following BA.1 infection [12].
- During the recent COVID-19 wave in the Republic of Korea, the Omicron variant was detected in most patients with COVID-19. Furthermore, the characteristics of the Omicron variant were compared with those of the S protein D614G mutant (detected in the GH clade predominant during the second and third waves) and Delta variant (2020–2021) in the Republic of Korea, focusing on its increased transmissibility. Specifically, epidemiological data collected at the time of diagnosis were used to compare viral load from the day of infection to 5 days post-infection. We aimed to provide sufficient data to inform effective responses to the transmission of the Omicron variant.
Introduction
- Specimen Collection and Real-Time Reverse-Transcription Polymerase Chain Reaction Testing for SARS-CoV-2
- In total, 19,996 COVID-19-positive specimens (nasopharyngeal and oropharyngeal swabs) were collected. Among them, 17,086 specimens were collected from infected patients in the second and third waves in 2020, 1,691 were from patients infected with the Delta variant in 2021, and 1,219 were from patients infected with the BA.1.1 (1,084 cases) and BA.2 (135 cases) sublineages of the Omicron variant. The specimens were subjected to RNA extraction, followed by real-time reverse-transcription polymerase chain reaction (RT-PCR) [13]. Briefly, RNA was extracted from 140 µL of each sample using a Viral RNA Mini Kit (Qiagen), according to the manufacturer’s protocol. Subsequently, RT-PCR was performed and the cycle threshold (Ct) value of the SARS-CoV-2 target genes was determined. The primer and probe sequences used for RNA-dependent RNA polymerase gene detection were as follows: 5′-GTGARATGGTCATGTGTGGCGG-3′ (forward), 5′-CARATGTTAAASACACTATTAGCATA-3′ (reverse), and 5′-CAGGTGGAACCTCATCAGGAGATGC-3′ (probe in 5-FAM/3′-BHQ format). The primer and probe sequences used for E gene detection were as follows: 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′ (forward), 5′-ATATTGCAGCAGTACGCACACA-3′ (reverse), and 5′-ACACTAGCCATCCTTACTGCGCTTCG-3′ (probe in 5-FAM/3′-BHQ format). All specimens were handled in a biosafety cabinet, according to the laboratory biosafety guidelines from the Korea Disease Control and Prevention Agency for COVID-19.
- Lineage Assignment of SARS-CoV-2 by Full-Genome and S Protein Sequencing
- This study analyzed more than 1,600 cases (random sampling) per week, with reference to European Centre for Disease Control recommendations to detect mutations that may exist at a rate of 1% in 100,000 confirmed COVID-19 cases per week.
- For full-genome sequencing, cDNA was amplified from the total RNA using ARTIC primer pools (https://artic.network/ncov-2019), QIAseq SARS-CoV-2 Primer Panel, and QIAseq FX DNA Library UDI Kit (Qiagen). Libraries were prepared using the Nextera DNA Flex Library Prep Kit (Illumina). Sequencing was performed on a MiSeq instrument using MiSeq Reagent Kit v2 (Illumina) to obtain an average genome coverage of >1,000× for all the samples. The reads were trimmed and mapped to the Wuhan-Hu-1 reference genome (GenBank accession number: MN908947.3) using CLC Genomics Workbench version 20.0.3 (CLC Bio) [14]. The lineages and clades of the SARS-CoV-2 sequences were assigned using Nextclade v1.7. [15] and PANGOLIN [16].
- The SARS-CoV-2 S protein-encoding gene was amplified using 1-step RT-PCR (Qiagen) with 6 primers selected from the ARTIC primer pools. Three overlapping fragments were amplified, purified, and sequenced at BIOFACT Co., Ltd.
- Calculation of Viral Copy Numbers
- Plasmids carrying the SARS-CoV-2 E gene were used as a positive control. A standard curve was constructed based on the plasmid concentrations. The regression equation (y=–3.5705x+39.055; y, Ct; x, copy) was obtained using the standard curve. The viral copy number in each sample was calculated using the above equation and expressed as a logarithm of base 10 [17].
- Statistical Analysis
- Statistical analysis was performed using SAS ver. 9.4 (SAS Inc.). The frequency and percentage of categorical variables were obtained, and the median and interquartile range were calculated for the Ct values and age. One-way analysis of variance was used to examine differences in the Ct values for each variable. The threshold for statistical significance was set at less than 0.05 in all cases [16].
- Ethics Approval
- All procedures involving human participants were performed in accordance with the ethical standards of the institutional and/or national research committees and the 1964 Declaration of Helsinki, including its later amendments, or with comparable ethical standards. The study was approved by the institutional review board of the Korea Disease Control and Prevention Agency (approval number: 2020-03-01-P-A) and was designated as a service to public health during the pandemic. Therefore, the review board waived the requirement for written informed consent from the participants, as outlined in the Title Laboratory Respondence to COVID-19.
Materials and Methods
- Characterization of the Confirmed Cases
- After the first report of COVID-19 in mid-January 2020, the S, L, and V clades occurred predominantly among infections in the Republic of Korea until March 2020. Subsequently, the G, GH, and GR clades were reported and have been predominant globally since April 2020, with an increased mutation frequency in the S protein-encoding gene. These mutations affected virological features including viral transmission and disease severity. The S protein D614G mutant, identified in the GH clade, was predominant until December 2020; the Delta variant of the GK clade was detected between April and July 2021. The latest wave, caused by the highly transmissible Omicron variant, is ongoing (Table 1).
- We analyzed 17,086 cases of D614G mutant infection reported in 2020, 1,691 cases of Delta variant infection, and 1,219 cases of infection with the BA.1.1 (1,084 cases) and BA.2 (135 cases) sublineages of the Omicron variant during the recent wave. Data was taken within 5 days of symptom onset. The clinical symptoms ranged from no symptoms to mild symptoms (fever, cough, and sore throat). The highest infection rate (32.1%) was observed in patients above the age of 60 and was caused by the D614G mutant. During the Delta wave, the highest infection rate (27.7%) was observed in patients aged 20–29 years. The infection rates of the BA.1.1 and BA.2 sublineages in the same age group were 22.5% and 24.4%, respectively. The overall incidence of COVID-19 was higher in women (51.6%) than in men (48.4%). However, the incidence of Delta variant infection was higher among men (52.6%). The Ct distribution of the SARS-CoV-2 strain isolated during the D614G wave showed the highest rate of infection (25.8%) in the 15–20-year age group; a similar Ct distribution was observed for the Delta variant, which also showed the highest rate of infection (42.6%) in that age group. The Ct value of individuals with the highest infection rate from the BA.1.1 and BA.2 sublineages was relatively low in patients below 15 years of age (62.0% and 83.7%, respectively); however, the value was higher than that from the D614G mutant and Delta variant (Table 2). In addition, statistical significance (p<0.001) was confirmed in the age and sex of infected individuals and the Ct of the D614G mutant, Delta, Omicron BA.1.1, and BA.2 variants.
- Daily Average Ct and Viral Load after Symptom Onset
- The overall mean Ct value within 5 days of symptom onset was 21.41 for the D614G mutant (range, 7.06–38.65; 95% confidence interval [CI], 21.31–21.51), 16.43 for the Delta variant (range, 7.61–34.40; 95% CI, 16.22–16.64), 15.36 for the BA.1.1 sublineage (range, 9.55–31.78; 95% CI, 15.15–15.57), and 13.58 for the BA.2 sublineage (range, 10.90–24.54; 95% CI, 13.25–13.91). Notably, the Ct value for the Omicron sublineages was lower than those for the D614G mutant and Delta variant. For the latter 2 variants, the Ct value gradually decreased each day after symptom onset. Conversely, the Ct value of the BA.1.1 and BA.2 sublineages hardly changed (Table 3; Figure S1). In addition, statistical significance (p<0.0001) was confirmed among each variant group and day. For the BA.2 sublineage, a high viral load was seen soon after symptom onset. All Ct values were measured using RT-PCR reagents and equipment, based on the same primers and probes. However, since the data were collected from different laboratories, variations may have occurred, as virus extraction protocols and RT-PCR performance may differ in different laboratory conditions, including operator and extraction methods. Therefore, a quality evaluation of the diagnostic laboratories was conducted to minimize the potential for detection bias.
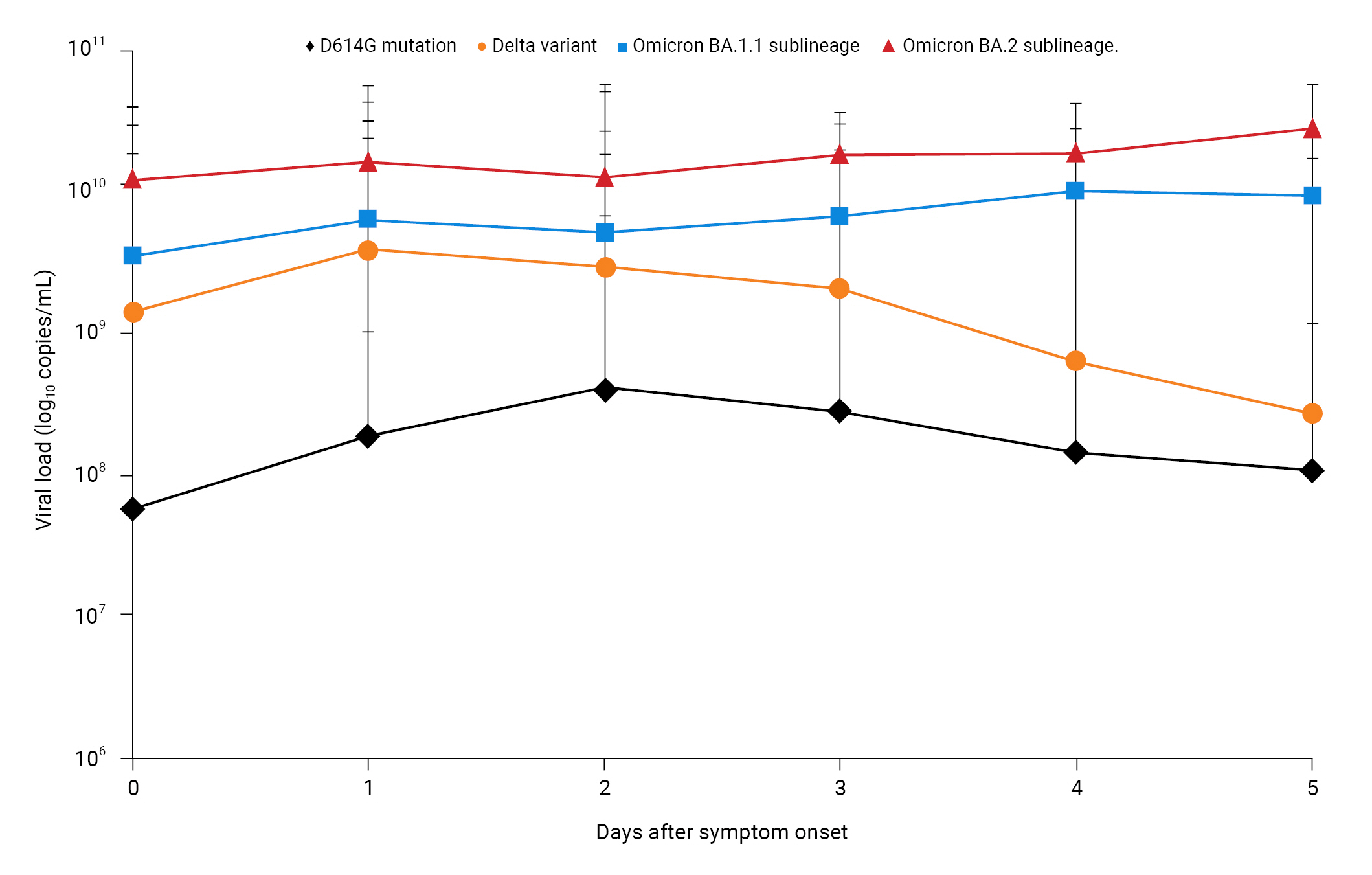
- The Ct value is inversely proportional to the amount of target nucleic acid in a sample. To compare the differences in viral load, the mean Ct value within 5 days of symptom onset was converted to viral load. Patients infected with the BA.2 sublineage had an approximately 155-fold higher viral load (1.36×1010 copies/mL) than those with the D614G mutant (8.76×107 copies/mL), a 6-fold higher viral load than those with the Delta variant (2.16×109 copies/mL), and a 3-fold higher viral load (4.32×109 copies/mL) than those with the BA.1.1 sublineage. Furthermore, patients infected with BA.2 had a viral load of 1.22×1010 copies/mL on the day of symptom onset (day 0); this was approximately 214-fold higher than that in patients with the D614G mutant (5.69×107 copies/mL), 9-fold higher than that in patients with the Delta variant (1.42×109 copies/mL), and 4-fold higher than that in patients with the BA.1.1 sublineage (3.44×109 copies/mL). Similarly, the viral load on day 4 after symptom onset (1.84×1010 copies/mL) was approximately 131-fold higher than that in patients with the D614G mutant (1.41×108 copies/mL), 30-fold higher than that in patients with the Delta variant (6.17×108 copies/mL), and 2-fold higher than that in patients with the BA.1.1 sublineage (9.79×109 copies/mL). Notably, within 4 days of symptom onset, the viral load was considerably higher for the BA.2 sublineage of Omicron than for the D614G mutant (approximately 32–214-fold), Delta variant (approximately 4–30-fold), and BA.1.1 sublineage (approximately 2–4-fold). This suggests that the BA.2 sublineage is associated with a high viral load at disease onset (Figure 1).
Results
- In the Republic of Korea, SARS-CoV-2 viral load was investigated as a potential contributing factor to the rise in Omicron infections observed in early 2022 compared with those seen until the end of 2021. The initial viral load within 5 days of symptom onset was found to be approximately 32–214-fold higher from the BA.2 sublineage than that observed during the D614G wave (mainly caused by the GH clade). Furthermore, BA.2’s viral load was approximately 4- to 30-fold and 2- to 4-fold higher than from the Delta variant and BA.1.1 sublineage, respectively. Additionally, previous studies have indicated that BA.2 is inherently more transmissible than BA.1 [12]. However, this difference in transmissibility is considerably smaller than that between BA.1 and Delta [11,18]. As of the time of writing for this manuscript, according to the Global Initiative on Sharing All Influenza Data, the incidence of BA.2 is proportionally increasing to other Omicron sublineages (BA.1 and BA.1.1) [7]. Thus, patients infected with the BA.1.1 and BA.2 sublineages of the Omicron variant may show an increased viral transmission. This may be an important factor contributing to the increased number of patients with COVID-19 during the Omicron wave (mean, >100,000 per day) compared with that observed during the D614G (mean, 186 per day) and Delta waves (mean, 713 per day). The current data suggest high viral replication and transmission; thus, epidemiological studies are required to determine the reproduction rate of this variant.
- The SARS-CoV-2 S protein interacts with the host’s angiotensin-converting enzyme 2 (ACE2) receptor through its receptor-binding domain [19,20]. However, a mutation in the receptor-binding domain can directly affect the interaction between the virus and ACE2. SARS-CoV-2, which harbors the D614G mutation, in which glutamic acid (D) at 614 is substituted with glycine (G) in the S protein, exhibits an improved ability to bind to ACE2, further enabling it to infect humans and cause widespread transmission [9,21]. The H69/V70 deletion is also associated with a 2-fold increase in infectivity, whereas the N501Y and K417N counterparts are associated with stronger binding to ACE2 [10]. Additionally, the P681H mutation in the furin cleavage domain at 681 to 687 prevents the proper formation of the S1/S2 unit, thereby altering the infectivity and pathogenicity of the virus [22,23]. The Omicron variant harbors a mutation at 614 in the S protein receptor-binding domain, as well as K417N, T478K, N501Y, and P681H mutations in the furin cleavage site of the S protein [13]. The increased transmission of the Omicron variant has been attributed to the D614G and H69/V70 deletions, as well as the K417N, T478K, and N501Y mutations. The P681H mutation is associated with altered viral infectivity and pathogenicity.
- Our results suggest that the increased viral load of the Omicron variant may have influenced the scale of the recent wave. However, additional studies are required to determine the association between clinical disease severity and virological features. A previous study reported increased transmissibility compared with that of the S protein D614G mutant and the Alpha and Delta variants, which was related to conformational stability (i.e., binding free energy between the S protein and ACE2) [24]. Therefore, further research should be performed to compare the trend of disease severity caused by the high viral loads of the BA.1.1 and BA.2 sublineages of the Omicron variant with previous waves and analyze vaccine efficacy in light of the current Omicron wave.
Discussion
- • The viral loads of Omicron variant from 5 days after symptom onset were compared with the Delta variant and the spike protein D614G mutant.
- • The Omicron variant exhibited a higher viral load than other variants, resulting in greater transmissibility within 5 days of symptom onset.
- • Future research should focus on vaccine efficacy against the Omicron variant and compare trends in disease severity associated with its high viral load.
HIGHLIGHTS
Supplementary Material
Figure S1.
-
Ethics Approval
All procedures involving human participants were performed in accordance with the ethical standards of the institutional and/or national research committees and the 1964 Declaration of Helsinki, including its later amendments, or with comparable ethical standards. The study was approved by the institutional review board of the Korea Disease Control and Prevention Agency (approval number: 2020-03-01-P-A) and was designated as a service to public health during the pandemic. Therefore, the review board waived the requirement for written informed consent from the participants, as outlined in the Title Laboratory Respondence to COVID-19.
-
Conflicts of Interest
The authors have no conflicts of interest to declare.
-
Funding
This work was supported by the Korea Disease Control and Prevention Agency (grant number: 6300-6331-301).
-
Availability of Data
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request. The SARS-CoV-2 whole genome and Sanger sequences are available in the Global Initiative on Sharing All Influenza Data (http://www.gisaid.org/).
-
Authors’ Contributions
Conceptualization: JMK, EJK; Data curation: JMK; Formal analysis: JMK, DK; Funding acquisition: CKY, EJK; Investigation: JMK, NJL, SHW; Methodology: JMK, DK, EJK; Project administration: CKY, EJK; Software: JMK; Resources: NJL, SHW, JL, HL, AKP, JAK, CYL, IHK; Supervision: CKY, EJK; Validation: JMK, EJK; Visualization: JMK; Writing–original draft: all authors; Writing–review & editing: all authors. All authors read and approved the final manuscript.
-
Additional Contributions
The authors thank all individuals who assisted in the collection and transport of patient specimens, the staff at the Korea Disease Control and Prevention Agency and local government, and private testing agencies for their contributions to controlling COVID-19.
Article information

- 1. World Health Organization (WHO). COVID-19 weekly epidemiological update, edition 92, 18 May 2022 [Internet]. WHO; 2022 [cited 2023 Jun 21]. Available from: https://apps.who.int/iris/handle/10665/354476.
- 2. Ong SW, Chiew CJ, Ang LW, et al. Clinical and virological features of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.351 (Beta), and B.1.617.2 (Delta). Clin Infect Dis 2022;75:e1128−36.ArticlePubMedPMCPDF
- 3. World Health Organization (WHO). Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern [Internet]. WHO; 2021 [cited 2023 Jun 21]. Available from: https://who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern.
- 4. Viana R, Moyo S, Amoako DG, et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022;603:679−86.PubMedPMC
- 5. Kim JM, Kim D, Rhee JE, et al. Isolation of the Omicron variant of SARS-CoV-2. Public Health Wkly Rep 2022;15:2−4.
- 6. World Health Organization (WHO). Coronavirus disease (COVID-19) pandemic [Internet]. WHO; 2023 [cited 2023 Jun 21]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- 7. GISAID [Internet]. GISAID; 2023 [cited 2023 Jun 21]. Available from: https://www.gisaid.org.
- 8. Outbreak.info. Omicron variant report [Internet]. Scripps Research; 2023 [cited 2023 Jun 21]. Available from: https://outbreak.info/situation-reports/omicron?loc=ZAF&loc=GBR&loc=USA&selected=ZAF.
- 9. Ozono S, Zhang Y, Ode H, et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun 2021;12:848. ArticlePubMedPMCPDF
- 10. Ramesh S, Govindarajulu M, Parise RS, et al. Emerging SARS-CoV-2 variants: a review of its mutations, its implications and vaccine efficacy. Vaccines (Basel) 2021;9:1195. ArticlePubMedPMC
- 11. World Health Organization (WHO). Statement on Omicron sublineage BA.2 [Internet]. WHO; 2022 [cited 2023 Jun 21]. Available from: https://www.who.int/news/item/22-02-2022-statement-on-omicron-sublineage-ba.2.
- 12. Yamasoba D, Kimura I, Nasser H, et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell 2022;185:2103−15.PubMedPMC
- 13. Kim JM, Chung YS, Jo HJ, et al. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res Perspect 2020;11:3−7.ArticlePubMedPMCPDF
- 14. Park AK, Kim IH, Kim J, et al. Genomic surveillance of SARS-CoV-2: distribution of clades in the Republic of Korea in 2020. Osong Public Health Res Perspect 2021;12:37−43.ArticlePubMedPMCPDF
- 15. Nextstrain Project [Internet]. Nextstrain; 2023 [updated 2023 Jun 18; cited 2023 Jun 21]. Available from: https://nextstrain.org/ncov/gisaid/global.
- 16. Rambaut A, Holmes EC, O'Toole A, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020;5:1403−7.ArticlePubMedPMCPDF
- 17. Kim JM, Rhee JE, Yoo M, et al. Increase in viral load in patients with SARS-CoV-2 Delta variant infection in the Republic of Korea. Front Microbiol 2022;13:819745. ArticlePubMedPMC
- 18. Lyngse FP, Kirkeby CT, Denwood M, et al. Household transmission of SARS-CoV-2 Omicron variant of concern subvariants BA.1 and BA.2 in Denmark. Nat Commun 2022;13:5760. ArticlePubMedPMCPDF
- 19. Bahrami A, Ferns GA. Genetic and pathogenic characterization of SARS-CoV-2: a review. Future Virol 2020;15:533−49.Article
- 20. Deshpande A, Harris BD, Martinez-Sobrido L, et al. Epitope classification and RBD binding properties of neutralizing antibodies against SARS-CoV-2 variants of concern. Front Immunol 2021;12:691715. ArticlePubMedPMC
- 21. Volz E, Hill V, McCrone JT, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 2021;184:64−75.ArticlePubMedPMC
- 22. Lubinski B, Fernandes MH, Frazier L, et al. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience 2022;25:103589. ArticlePubMedPMC
- 23. Johnson BA, Xie X, Bailey AL, et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 2021;591:293−9.ArticlePubMedPMCPDF
- 24. Choi KE, Kim JM, Rhee JE, et al. Molecular dynamics studies on the structural stability prediction of SARS-CoV-2 variants including multiple mutants. Int J Mol Sci 2022;23:4956. ArticlePubMedPMC
References
Figure & Data
References
Citations




 Cite
Cite