Detection and Isolation of SARS-CoV-2 in Serum, Urine, and Stool Specimens of COVID-19 Patients from the Republic of Korea
Article information
Abstract
Objectives
Coronavirus Disease-19 (COVID-19) is a respiratory infection characterized by the main symptoms of pneumonia and fever. It is caused by the novel coronavirus severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2), which is known to spread via respiratory droplets. We aimed to determine the rate and likelihood of SARS-CoV-2 transmission from COVID-19 patients through non-respiratory routes.
Methods
Serum, urine, and stool samples were collected from 74 hospitalized patients diagnosed with COVID-19 based on the detection of SARS-CoV-2 in respiratory samples. The SARS-CoV-2 RNA genome was extracted from each specimen and real-time reverse transcription polymerase chain reaction performed. CaCo-2 cells were inoculated with the specimens containing the SARS-COV-2 genome, and subcultured for virus isolation. After culturing, viral replication in the cell supernatant was assessed.
Results
Of the samples collected from 74 COVID-19 patients, SARS-CoV-2 was detected in 15 serum, urine, or stool samples. The virus detection rate in the serum, urine, and stool samples were 2.8% (9/323), 0.8% (2/247), and 10.1% (13/129), and the mean viral load was 1,210 ± 1,861, 79 ± 30, and 3,176 ± 7,208 copy/µL, respectively. However, the SARS-CoV-2 was not isolated by the culture method from the samples that tested positive for the SARS-CoV-2 gene.
Conclusion
While the virus remained detectable in the respiratory samples of COVID-19 patients for several days after hospitalization, its detection in the serum, urine, and stool samples was intermittent. Since the virus could not be isolated from the SARS-COV-2-positive samples, the risk of viral transmission via stool and urine is expected to be low.
Introduction
The Coronavirus Disease-19 (COVID-19) leads to the development of Severe Acute Respiratory Syndrome (SARS), with a fever, cough, sore throat, myalgia, and headache as the most common clinical symptoms. It is a novel viral infection that first emerged in Wuhan, China in December 2019 as a pneumonia of unknown etiology [1-4]. Coronaviruses infect humans and various animals, and subsequently cause respiratory, enteric, hepatic, and neurologic diseases [5,6]. Most human coronaviruses, apart from the SARS-CoV and Middle East Respiratory Syndrome (MERS)-CoV, infect the upper respiratory tract and lead to the development of mild respiratory symptoms with a low fatality rate. Conversely, the SARS-CoV and MERS-CoV mostly infect the lower respiratory tract and may lead to the development of severe respiratory symptoms, including shortness of breath and pneumonia.
The novel SARS-CoV-2, the etiologic agent of COVID-19, is a single-stranded enveloped RNA virus of the family Coronaviridae that includes 4 genera: alphacoronaviruses, betacoronaviruses, deltacoronaviruses, and gammacoronaviruses. Of the human coronaviruses (HCoV), HCoV-229E and HCoV-NL3 are alphacoronaviruses, while HCoV-OC43 and HCoV-HKU1 are betacoronaviruses [3,4]. The SARS-CoV and the MERS-CoV, which first emerged in 2002 and 2012, respectively, belong to the betacoronavirus genera [1,7]. The SARS-CoV-2 is known to have a higher transmission rate and infectivity than SARS-CoV and MERS-CoV [7-9].
Presence of a fever and a cough are the primary clinical manifestations of COVID-19 however, patients also exhibited other symptoms such as nausea, vomiting, diarrhea, and abdominal discomfort [3]. Presently, the most common method used for COVID-19 diagnosis is the detection of SARS-CoV-2 in upper and lower respiratory specimens, including nasopharyngeal swabs, oropharyngeal swabs, sputum, lower respiratory tract aspirates, and bronchoalveolar lavage. Genetic testing methods, such as real-time reverse transcription polymerase chain reaction (RT-PCR), are the standard methods of laboratory testing for COVID-19 that are currently in use in most countries.
In the present study, we investigated whether SARS-CoV-2, which infects humans and may subsequently trigger the development of various clinical symptoms, can be detected in body fluids such as serum, urine, and stool, besides respiratory specimens. In addition, we aimed to isolate the virus from SARS-CoV-2-positive samples to determine viral infectivity.
Materials and Methods
1. Specimens from COVID-19 patients
To examine viral shedding in body parts other than the respiratory tract, and the infectivity of the detected virus, respiratory specimens such as nasopharyngeal swab, oropharyngeal swab or sputum, as well as serum, urine, and stool specimens were non-periodically sampled from 74 COVID-19 patients admitted in a hospital between January 19th and March 30th, 2020.
Of the respiratory specimens, the upper respiratory samples were collected at least twice from all patients. Serum, urine, and stool samples were collected from 71, 54, and 38 patients, respectively. Each sample had been tested by the Korea Centers for Diseases Control and Prevention to monitor the SARS-CoV-2 infection status of the COVID-19 patients.
2. RNA extraction and real-time RT-PCR
Serum samples were collected in a serum separation tube and centrifuged. Urine samples were centrifuged and the supernatants removed. The pellet was resuspended in 1-2 mL serum-free Dulbecco’s Modified Eagle Medium (DMEM). Each stool sample (1 g) was suspended in 10 mL of phosphate-buffered saline and was centrifuged to collect the supernatant for RNA extraction. RNA extraction and real-time RT-PCR was performed on the samples according to the method proposed by Kim et al in 2020 [10]. Briefly, RNA was extracted from 140 µL of the sample using a Qiagen viral RNA mini kit (Qiagen, Hilden, Germany) according to the method recommended by the manufacturer. Real-time RT-PCR was performed with the extracted RNA, and the cycle threshold value of the SARS-CoV-2 target gene was determined.
3. Virus isolation
To isolate SARS-CoV-2 from samples that tested positive for the virus in real-time RT-PCR analyses, the samples were mixed with a 1:1 nystatin (10,000 U/mL) and penicillin-streptomycin (10,000 U/mL) mixture in a 1:4 ratio, and left to react at 4°C for 1 hour. The samples were then centrifuged at 400× g for 10 minutes and the supernatant was used as the inoculant. For the cell inoculation, cells were cultured from the CaCo-2 cell line (derived from human epithelial colorectal adenocarcinoma cells) in DMEM supplemented with 20% fetal bovine serum and 1% penicillin, and were incubated at 37°C, 5% CO2. On the day prior to inoculation, the cells were seeded at 2 × 105 cells/well into a 12-well plate. On the day of the primary inoculation, each well was replaced with DMEM supplemented with 2% fetal bovine serum, and 100 µL of the sample was used for the inoculation. The cells were cultured for 5 days and then harvested. The cell culture supernatant was centrifuged at 2,000 rpm for 10 minutes and the supernatant collected. The same method was repeated for a secondary inoculation. Five days after the secondary inoculation, the cell culture supernatant was harvested, centrifuged at 600 × g for 10 minutes, and the supernatant collected. To evaluate the viral replication process, RNA from the secondary inoculation cell culture supernatant was extracted and assessed for the presence of SARS-CoV-2 using real-time RT-PCR. The virus isolation experiment was conducted in a biosafety Level-3 facility.
4. Calculation of the viral copy number
According to the standard curve, the plasmid DNA (E and RdRp genes) containing the linearized form of the PCR product (E gene, 113 bp; RdRp gene, 100 bp) were inserted into the pGEM-T Easy vector system by TA Cloning, followed by the transformation of DH10B cells (Promega Corporation, WI, USA), and the purification of plasmids using the QIAprep Spin Miniprep Kit and Qiagen Plasmid Plus Midikit (Qiagen, Hilden, Germany). The plasmid concentration was determined by measuring optical density with a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, USA), and the number of viral genome equivalent copies was calculated using the equation: amount of DNA (E gene: 7.4 × 1,011 copy/µL, RdRp gene: 8.4 × 1,011 copy/µL).
5. Ethics statement
The study was approved by the Institutional Review Board at the Korea Centers for Disease Control and Prevention (2020-03-01-P-A). The board waived the requirement for written consent.
Results and Discussion
The presence of SARS-CoV-2 was tested in serum, urine, and stool samples collected from 74 COVID-19 patients, using real-time RT-PCR. Of the patients, 44 were male and 30 were female aged 9-80 years (median 43 years). The maximum period of sample collection was 17 days from the date of disease confirmation. From the 74 patients, 323 serum samples, 247 urine samples, and 129 stool samples were collected and tested. The SARS-CoV-2 RNA detection rates were 2.8% (9/323), 0.8% (2/247), and 10.1% (13/129), respectively. In 15 patients (20.0%), SARS-CoV-2 was detected at least once in at least 1 of the 3 specimens, apart from the respiratory specimens (Figure 1). In 4 of these patients, the virus was detectable in the serum and stool samples for a longer period than in the respiratory samples. The virus was detected in serum samples of 6 patients, urine samples of 2 patients, and stool samples of 8 patients.
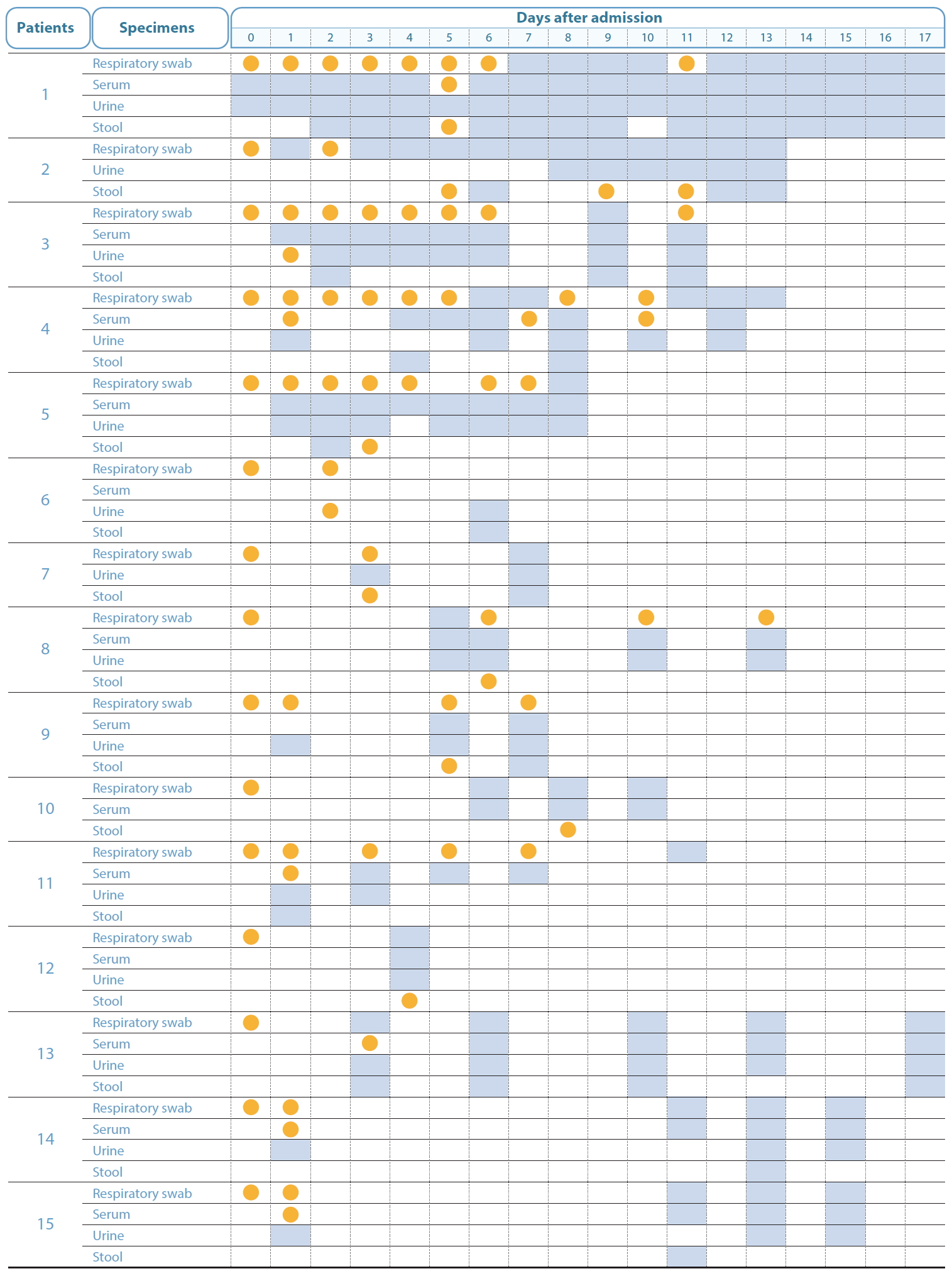
The detection of SARS-CoV-2 RNA in respiratory swabs, serum, urine, and stool samples in the days following disease confirmation, during the course of infection in COVID-19 patients. Orange represents the detection of viral RNA, grey represents undetected viral RNA, and white indicates that the samples were not collected, and therefore, not tested.
The number of viral genome equivalent copies in the serum (19-6, 127 copies/µL), urine (49-109 copies/µL), and stool samples (27-27, 310 copies/µL), were calculated on the basis of the RdRp gene copy number. In 3 patients, the virus was not detected in the respiratory samples, although it was present in stool samples; in 2 of these patients, the virus could be detected in the stool samples at Days 7 and 9, even after it was no longer detectable in the respiratory samples. To isolate the virus, CaCo-2 cells were inoculated with the SARS-CoV-2 positive serum, urine, or stool sample. After primary inoculation, the cells were cultured for 5 days and then a secondary inoculation was performed however, the virus could not be isolated from the samples.
There have been no published reports of SARS-CoV-2 viremia to date. The SARS-CoV-2 RNA level detected was 50% in plasma and 78% in serum samples at 7- or 14-days following hospitalization [11,12]. In other studies, the plasma viral load of SARS-CoV-2 peaked at 4-8 days after the disease onset, and followed a pattern of transient viremia, or occasionally persisted for more than 10 days, depending on the patient [13]. In this study, the viral RNA was detected in the serum of 5 patients at 3-6 days after onset. However, owing to irregular and discontinuous blood sampling, the duration for which the RNA was detectable could not be assessed (Figure 2). In addition, the inhibitory effect of the antiviral drug administered during treatment after hospitalization on viral replication cannot be excluded. Blood samples were collected 1-3 times within 6 days after onset in 46% of the patients. Since SARS-CoV-2 can be transmitted during the asymptomatic stage of infection before disease onset [14,15], future studies should investigate viremia during the asymptomatic stage and determine the rate of viremia.
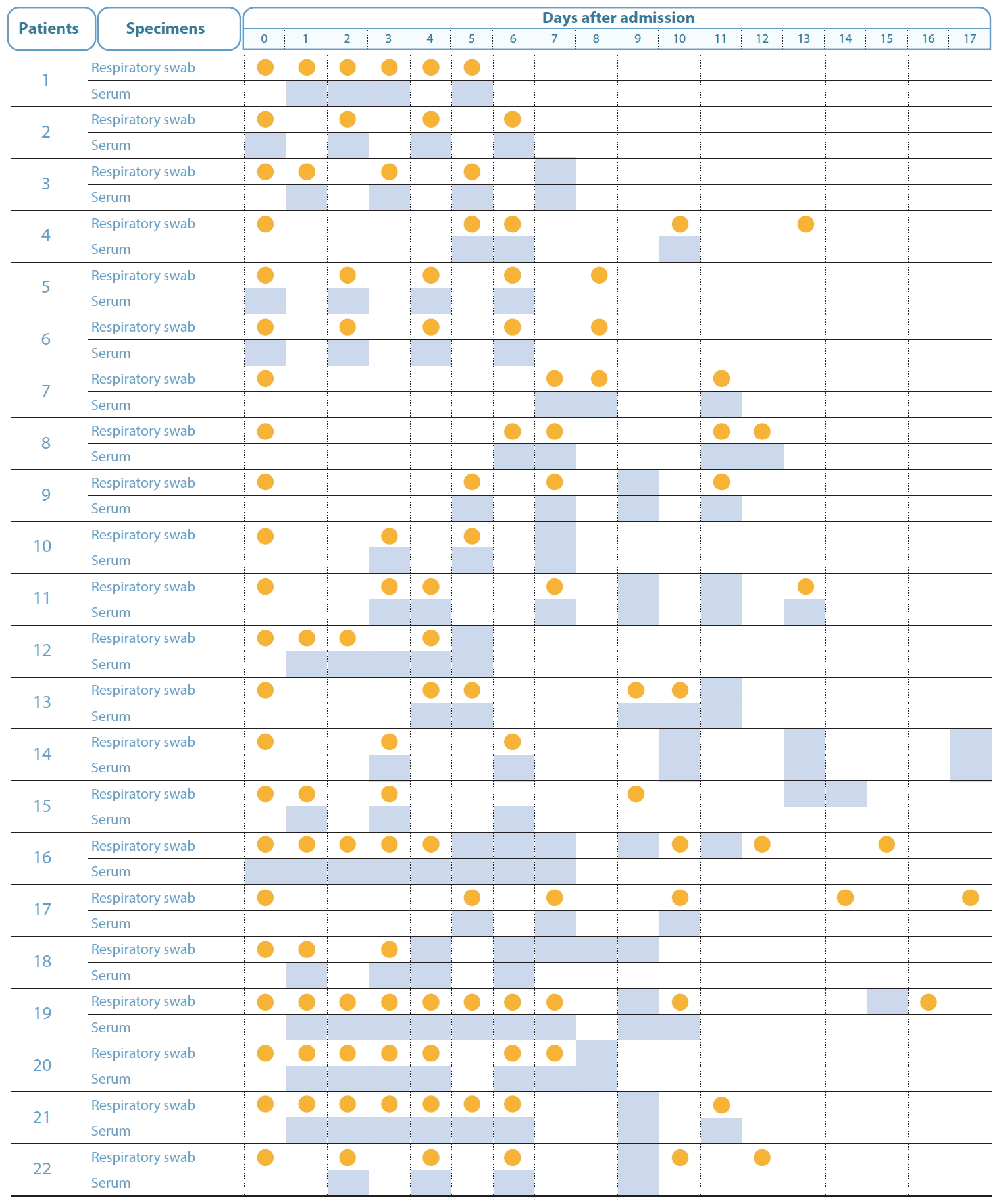
Time points for respiratory swab and serum sample collection after admission to hospital after SARS-CoV-2 RNA detection. Orange represents the detection of viral RNA, grey represents undetected viral RNA, and white indicates that the samples were not collected, and therefore, not tested.
It has been reported that in SARS patients, viral shedding was detected in respiratory and stool specimens for up to 52 and 126 days, respectively [16]. However, in MERS patients, viral shedding has rarely been studied [17]. In a study on viral shedding in 37 MERS patients, the MERS-CoV RNA was detected in 93% of the lower respiratory samples for up to 3 weeks after disease confirmation, and viral RNA was detected in serum, stool, and urine samples [18]. However, virus could not be isolated from the stool or serum samples, which had the highest RNA concentration [19]. It has also been reported that MERS-CoV RNA was detected in stool samples, with the highest detection rate at Days 4-11 after onset, and was detected even at 30 days after onset in some patients [18,19].
Although it has been reported that SARS-CoV-2 RNA was not detectable in the upper respiratory tract at 21 days on average after confirmation [14], there was a specific case wherein the duration of viral shedding was observed at 20 days and 37 days [20]. Apart from detection of SARS-CoV-2 RNA in the upper respiratory tract, it has been reported that SARS-CoV-2 RNA has been detected in stool samples. In a study on 28 COVID-19 patients, the most common clinical symptoms were a cough (28.6%), sore throat (28.6%), and fever (25.0%), while diarrhea (10.7%) was not a common symptom [3]. A study reported that 10% of the patients had diarrhea and experienced nausea 1-2 days before the onset of fever and respiratory symptoms however, there were no reports of viral RNA detection in the samples collected from these patients [18]. In another study, 11.6% of the hospitalized patients exhibited gastrointestinal symptoms at admittance and 49.5% exhibited such symptoms during their hospital stay, and depending on the presence of gastrointestinal symptoms, the viral RNA detection rates were 52.4% and 39.1%, respectively [21]. However, the study that reported the detection of SARS-CoV-2 RNA in stool samples did not investigate the infectivity of the detected viral RNA.
The WHO recognizes the detection of SARS-CoV-2 RNA in lower and upper respiratory samples such as nasal swab, throat swab, and sputum as the official diagnostic method for COVID-19 testing. However, in addition to respiratory samples, viral RNA was also detected in serum, urine, or stool samples, suggesting the possibility of SARS-CoV-2 transmission via such specimens [22-25]. In this current study, SARS-CoV-2 RNA was detected in respiratory swabs as well as in serum, urine, and stool samples. Moreover, the virus from infected CaCo-2 cells was isolated to determine the infectivity of the viral RNA however, the presence of SARS-CoV-2 could not be confirmed in RNA-positive specimens, and the viral load was also considerably low. These results suggest that the SARS-CoV-2 is predominantly transmitted via the respiratory tract, and transmission via serum, urine, and stools is considerably limited.
Notes
Conflicts of Interest
The authors have no conflicts of interest to declare.
Acknowledgements
The authors thank all those who assisted in the collection and transport of patient samples. This study was funded by Korea Centers for Disease Control and Prevention (no.: 4800-4837-301).
