Results of contact tracing for SARS-CoV-2 Omicron sub-lineages (BA.4, BA.5, BA.2.75) and the household secondary attack risk
Article information
Abstract
Objectives
This study aimed to assess the contact tracing outcomes of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron sub-lineages BA.4, BA.5, and BA.2.75 within Republic of Korea, and to generate foundational data for responding to future novel variants.
Methods
We conducted investigations and contact tracing for 79 confirmed BA.4 cases, 396 confirmed BA.5 cases, and 152 confirmed BA.2.75 cases. These cases were identified through random sampling of both domestically confirmed and imported cases, with the goal of evaluating the pattern of occurrence and transmissibility.
Results
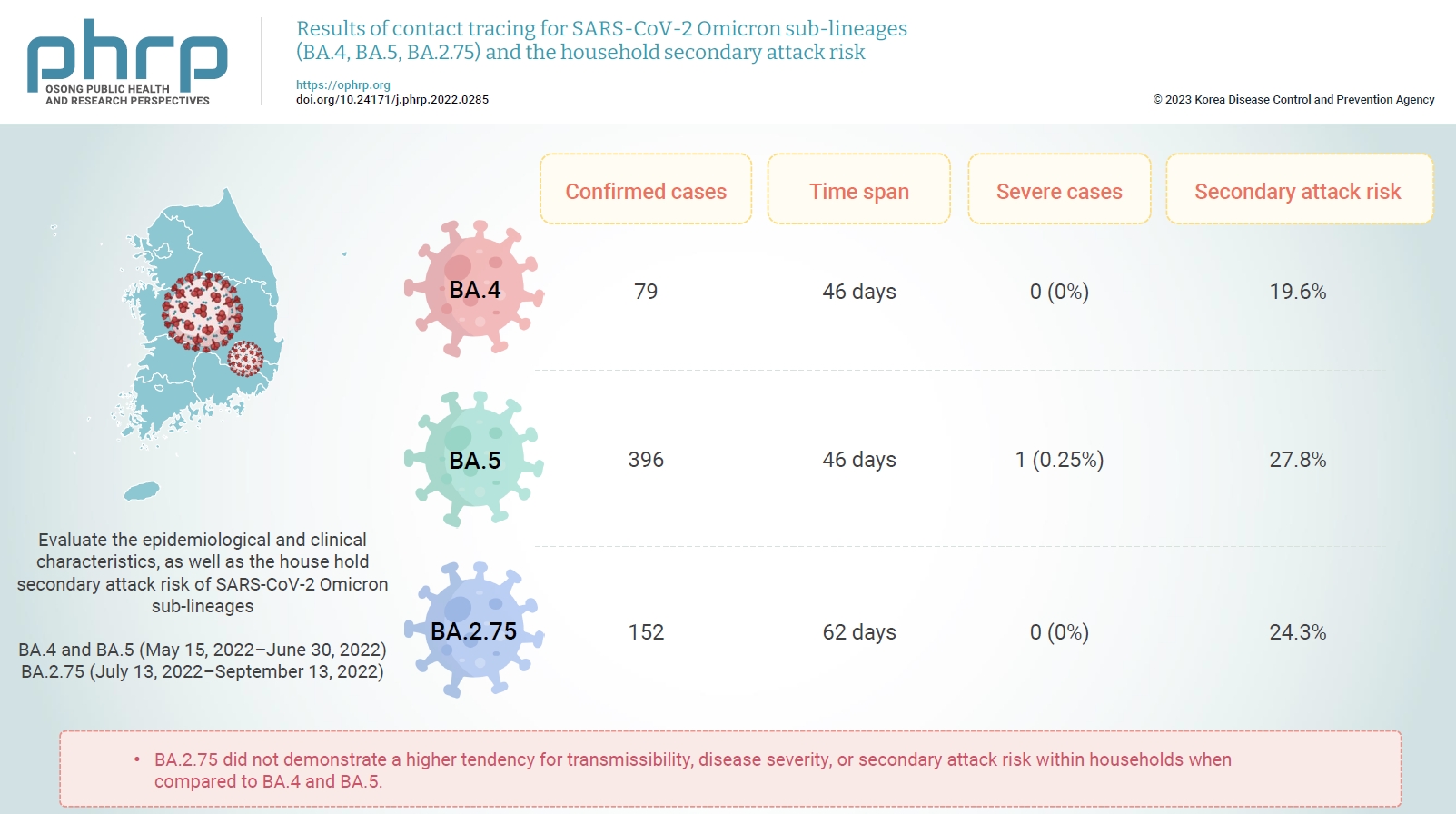
We detected 79 instances of Omicron sub-lineage BA.4 across a span of 46 days, 396 instances of Omicron sub-lineage BA.5 in 46 days, and 152 instances of Omicron sub-lineage BA.2.75 over 62 days. One patient with severe illness was confirmed among the BA.5 cases; however, there were no reports of severe illness in the confirmed BA.4 and BA.2.75 cases. The secondary attack risk among household contacts were 19.6% for BA.4, 27.8% for BA.5, and 24.3% for BA.2.75. No statistically significant difference was found between the Omicron sub-lineages.
Conclusion
BA.2.75 did not demonstrate a higher tendency for transmissibility, disease severity, or secondary attack risk within households when compared to BA.4 and BA.5. We will continue to monitor major SARS-CoV-2 variants, and we plan to enhance the disease control and response systems.
Introduction
Variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) represent a potential risk to global public health. As such, the World Health Organization is diligently tracking changes in these variants to classify them as either variants of interest (VOIs) or variants of concern (VOCs), underscoring the importance of genotype monitoring. By September 22, 2022, the SARS-CoV-2 Omicron variant B.1.1.529 had emerged as the dominant variant globally, making up over 98% of the viral sequences currently shared through the Global Initiative on Sharing All Influenza Data (GISAID) [1]. In the European Union, Omicron sub-lineages BA.2, BA.4, and BA.5 have been designated as VOCs due to their transmissibility and severity. However, the Omicron sub-lineage BA.2.75 is categorized and managed as a VOI due to inconclusive epidemiological and laboratory evidence [2]. A study of global occurrence trends for these major variants revealed 5,974 cases of BA.2.75 (prevalence, ≤0.5%) in 47 countries, 45,536 cases of BA.4 in 100 countries (prevalence, ≤0.5%), and 771,047 cases of BA.5 in 132 countries (prevalence, 6%) as of September 20, 2022 [3].
In late March 2022, South Korean disease control authorities implemented the coronavirus disease 2019 (COVID-19) Novel Variant Impact Assessment System to prepare for the domestic outbreak of newly emerging COVID-19 variants and to establish prompt responses to novel variants. Since the first report of the Omicron recombinant variant in Republic of Korea on April 12, 2022, major variant cases have been investigated, and risk assessments have been carried out [4,5]. These epidemiological investigations encompassed cases of variants identified by genome analysis, epidemiologically-linked cases, and contact tracing. If a variant is deemed dangerous based on the results of the epidemiological investigation and response, it is recommended to heighten the control standards [5].
The goal of this study was to compare and evaluate the epidemiological and clinical characteristics, as well as the household secondary attack risk, of confirmed BA.5, BA.4, and BA.2.75 cases, each of which was investigated individually. Moreover, we aimed to utilize these findings as foundational data for shaping and implementing future response policies to variant outbreaks.
Materials and Methods
Subjects
The subjects of this study were confirmed cases of BA.4 and BA.5 variants reported from May 15, 2022, when these variants were first identified in Republic of Korea, through June 30, 2022. The study also included confirmed cases of the BA.2.75 variant reported from July 13, 2022, the date when this variant was first discovered in Republic of Korea, through to September 13, 2022.
Definitions of Variables
Confirmed cases
A confirmed case of a COVID-19 Omicron sub-lineage was used to refer to an individual whose infection with a specific Omicron variant was confirmed through genomic analysis. Domestic and imported cases selected via random sampling were analyzed. The random sampling targeted respiratory patients, high-risk patients, and patients confirmed to have been infected overseas. Random sample targets were selected based on their likelihood of being a COVID-19 Omicron sub-lineage case, with the aim of identifying clinical and epidemiological characteristics.
Epidemiologically-linked cases
An epidemiologically-linked case was defined as a confirmed case that had contact with another confirmed case within a 2-week period before or after the onset of symptoms in the confirmed case. In essence, it refers to an individual who was confirmed to be infected through testing co-workers and family members who had contact during the transmissible period. However, cases involving other variants, identified through genomic analysis, were excluded.
Contacts
The term "contact" was used to refer to an unconfirmed individual who had contact with a confirmed case or an epidemiologically-linked case within a 2-week period before or after the onset of symptoms.
Household secondary attack risk
The household secondary attack risk was defined as the proportion of confirmed cases among household members who had contact with a confirmed (or primary) case, who had more than 1 household member, after they returned from overseas. This does not include household members who traveled overseas alongside the confirmed (or primary) cases. We calculated the exact binomial 95% confidence intervals and used the chi-square test to determine odds ratios.
Contact Tracing Methods
Investigations were carried out among household contacts who were registered in the COVID-19 information management system. These contacts were either self-reported by confirmed cases or reported through individual case investigations by local authorities. Polymerase chain reaction (PCR) testing for these household contacts was performed 3 times (on day 1, day 3, and day 9) within a 10-day period from the date of last exposure to enhance infection detection. However, for those individuals where 10 days had already passed since the last exposure, PCR testing was performed just once. Furthermore, no PCR testing was conducted for those individuals where 15 or more days had passed since the last exposure. Confirmed cases among household contacts were classified as epidemiologically-linked cases, and a genome analysis was conducted on available samples to ascertain the correlation with the respective confirmed cases. Confirmed cases in which another variant was identified were excluded from this study.
Ethics Approval
This study was granted an exemption from review by the Institutional Review Board (IRB) of the Korea Disease Control and Prevention Agency (IRB No: 2021-12-05-PE-A).
Results
General Demographic Characteristics
The monitoring results for the Omicron sub-lineages BA.4, BA.5, and BA.2.75 revealed that from May 15, 2022, to June 30, 2022, a span of 46 days since the initial confirmation of next-generation sequencing (NGS) analysis results, 97 confirmed cases of BA.4 and 396 confirmed cases of BA.5 were identified. Moreover, 152 confirmed cases of BA.2.75 were detected from July 13, 2022, to September 13, 2022, a period of 62 days since the initial NGS-based confirmation. When comparing the total number of confirmed cases and the occurrence status of confirmed cases by each Omicron sub-lineage during the monitoring period, it was observed that the total number of confirmed cases was decreasing, yet the counts of confirmed BA.4 and BA.5 cases were on the rise. Conversely, throughout the BA.2.75 monitoring period, the total number of confirmed cases consistently increased, though the incidence of BA.2.75 was lower compared to the other sub-lineages (Figure 1).
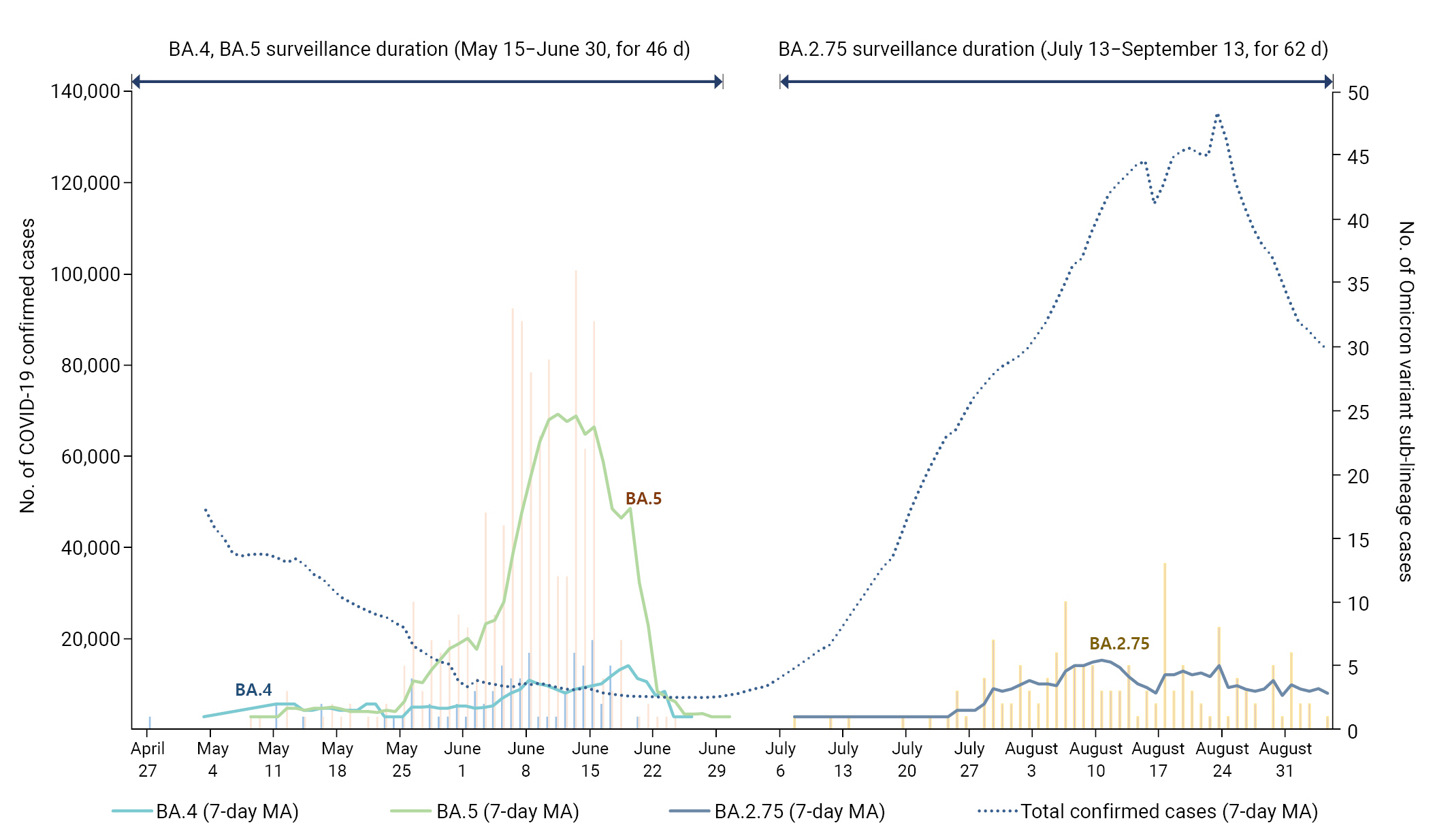
Incidence of cases during the monitoring period for severe acute respiratory syndrome coronavirus 2 Omicron variant sub-lineages (BA.4, BA.5, and BA.2.75). MA, moving average.
The epidemiological and clinical characteristics of confirmed cases for the Omicron sub-lineages BA.4, BA.5, and BA.2.75 were as follows: Men had a higher incidence than women across all 3 sub-lineages. The age group of 20 to 40 years constituted the majority of cases, metropolitan areas reported a higher incidence, very few cases had a history of prior infection, and most cases were mild. There were notable differences in the proportion of symptomatic cases and the route of infection. Most BA.4 and BA.2.75 cases were asymptomatic, and the majority of these cases were contracted overseas. For BA.5, most cases reported symptoms, and the majority were infected domestically (Table 1).

Epidemiological and clinical characteristics of SARS-CoV-2 Omicron variant sub-lineages (September 13, 2022)
The secondary attack risk among household contacts of confirmed Omicron sub-lineage cases (BA.4, BA.5, BA.2.75) was as follows: There were 33 BA.4 cases with household members, of which 3 were excluded due to having co-primary cases from overseas. This is why Table 2 shows 30 BA.4 primary cases. There were 193 BA.5 cases with household members, with 23 excluded because they had co-primary cases from overseas, resulting in 170 BA.5 primary cases in Table 2. Additionally, there were 72 BA.2.75 cases with household members, and 34 were excluded due to co-primary cases from overseas, leaving 38 BA.2.75 primary cases in Table 2. No statistically significant difference was found between the secondary attack risk of BA.2.75 and those of BA.4 and BA.5 (Table 2). In an analysis according to vaccination status, the secondary attack risk generally decreased with increasing vaccination of household contacts for BA.2.75 and BA.5 but not for BA.4 (Table 3).

SARS-CoV-2 infection attack risk among the household contacts of SARS-CoV-2 Omicron variant sub-lineages
Discussion
The epidemiological and clinical characteristics of BA.4, BA.5, and BA.2.75, which are the major VOCs among the Omicron variant sub-lineages, are described below.
In total, 79 cases of Omicron sub-lineage BA.4 and 396 cases of Omicron sub-lineage BA.5 were detected over a span of 46 days, while 152 cases of Omicron sub-lineage BA.2.75 were identified during a period of 62 days. Only 1 severe case was identified among BA.5 confirmed patients, while no severe cases were reported among the BA.4 and BA.2.75 patients. The secondary attack risk among household contacts was 19.6% for BA.4, 27.8% for BA.5, and 24.3% for BA.2.75. Compared to BA.4 and BA.5, BA.2.75 showed no increased tendency for transmissibility, disease severity, or household secondary attack risk. There were no statistically significant differences among the Omicron sub-lineages.
The number of confirmed cases of the 3 Omicron variant sub-lineages was higher in men than in women, predominantly affecting individuals in their 20s to 40s. These findings align with a United Kingdom (UK) study, which reported that most confirmed BA.2.75 cases affected patients aged 20 to 39 years, with a median age of 30 years [6]. This could be attributed to young people's active participation in activities such as overseas travel and business trips, potentially exposing them to SARS-CoV-2.
The occurrence of Omicron variant sub-lineages during the monitoring period identified 152 cases (detection rate as of September 10, 1.3%) of BA.2.75 over 62 days (July 13 to September 3), and 396 cases (detection rate as of July 2, 28.2%) of BA.5 over 46 days (May 15 to June 30) [7,8]. The BA.2.75 sub-lineage showed a slower rate of occurrence compared to the BA.5 sub-lineage, consistent with results from a UK study [2]. The term “occupancy rate” refers to the number of detected mutations over the same period. The BA.5 sub-lineage, due to its faster occupancy rate, spread rapidly in Republic of Korea following its introduction into the country, resulting in a higher percentage of domestic BA.5 cases compared to imported ones. Conversely, due to its slower occupancy rate, the percentage of imported BA.2.75 cases was higher than that of domestic cases.
Among the confirmed BA.5 cases, only 1 patient was identified as critically ill, with no critically ill patients or deaths reported among the confirmed BA.4 and BA.2.75 cases. These results support previous findings indicating the severity of the Omicron variant sub-lineages BA.2.75, BA.4, and BA.5 is not as high as that of the existing Omicron variants [9].
The secondary household attack risk for the Omicron sub-lineages BA.2.75, BA.4, and BA.5 were 24.3%, 19.6%, and 27.8%, respectively. Although the BA.5 sub-lineage, which is known for its high transmissibility, displayed the highest secondary attack risk, there was no statistically significant difference among the sub-lineages. Comparatively, a meta-analysis of 135 overseas studies reported a secondary attack risk of 42.7% among household contacts of confirmed Omicron variant cases. On the other hand, a previous study during the initial outbreak of the Omicron variant in Republic of Korea reported a rate of 65.0% [10,11]. The discrepancies between these study results can be attributed to various factors such as sample size, age, underlying diseases, and vaccination status of the study participants. This study also differed by excluding household contacts confirmed to have COVID-19 after entering the country with the primary cases, and only included household contacts infected via domestic transmission.
Additionally, the secondary attack rate might be overestimated because it is calculated based on the single-exposure assumption. In other words, estimates tend to inflate the secondary attack rate by assuming that all household contacts are infected by the primary case [12].
The investigation into the household secondary attack risk based on vaccination status revealed no significant difference in the secondary attack risk among household contacts, regardless of the index cases' vaccination status. However, an observed trend indicated a decrease in the secondary attack rate with an increase in vaccinations among household members of confirmed BA.5 and BA.2.75 cases. This result diverges from a previous study where the secondary attack risk among household contacts of unvaccinated index patients was higher than that with vaccinated index patients. The discrepancy between the 2 studies could have occurred because the previous study did not compare the household secondary attack risk based on the vaccination status of the household contacts, and the incidence rate may have been higher in index cases where more than 5 months had elapsed since vaccination than in unvaccinated index patients [13].
This study has certain limitations. Firstly, the monitoring period was short, resulting in a relatively small number of participants. Secondly, factors such as age, time elapsed since vaccination, and history of infections were not considered. Thirdly, foreign entrants were excluded from the study as it was not possible to ascertain their vaccination status. Lastly, when there was a possibility of simultaneous exposure abroad, household contacts were excluded from the analysis, potentially excluding some secondary cases.
Despite these limitations, the significance of this study lies in its identification of both the epidemiological and clinical characteristics of the less well-known Omicron sub-lineage BA.2.75, and the secondary attack risk among household contacts. Furthermore, this study demonstrated that the Omicron sub-lineage BA.2.75 did not have a higher rate of transmission than the existing BA.4 and BA.5 sub-lineages. Notably, our findings are significant as they represent the first comparative analysis of VOCs in Republic of Korea. The results of this study can guide disease control authorities to continue monitoring major SARS-CoV-2 variants in Republic of Korea, thereby continually refining and developing the disease prevention and response system.
HIGHLIGHTS
• Severe acute respiratory syndrome coronavirus 2 Omicron sub-lineages BA.2.75 showed slower accupancy rates than BA.4 and BA.5.
• One patient with severe illness was confirmed among the BA.5 cases, and there were no reports of severe illness in the confirmed BA.4 and BA.2.75 cases.
• The secondary attack risk among household contacts were no statistically significant difference. BA.2.75 did not demonstrate a higher tendency for transmissibility, disease severity, or secondary attack risk within households when compared to BA.4 and BA.5.
Notes
Ethics Approval
This study was exempted from review by the IRB of the Korea Disease Control and Prevention Agency (IRB No: 2021-12-05-PE-A).
Conflicts of Interest
The authors have no conflicts of interest to declare.
Funding
None.
Availability of Data
All data generated or analyzed during this study are included in this published article. For other data, if you have additional questions about the study, please contact the corresponding author (pahmun@korea.kr).
Authors’ Contributions
Formal analysis: MY, HYL, HJK; Software: SEL, YJS; Validation: JJ; Investigation: AKP, IHK; Supervision: YJP, EJK; Writing–original draft: MY; Writing–review & editing: all authors. All authors read and approved the final manuscript.
Acknowledgements
We would like to express our sincere appreciation to the epidemiological investigators at the regional response centers and those in charge of infectious diseases in local governments for their work and efforts in collecting data and on-site responding at front-line quarantine sites for this study.