The effect of photodynamic therapy using Radachlorin on biofilm-forming multidrug-resistant bacteria
Article information
Abstract
Objectives
This study aimed to test the effect of photodynamic therapy (PDT) on the inhibition and removal of biofilms containing multidrug-resistant Acinetobacter baumannii.
Methods
Using multidrug-resistant A. baumannii strains, an antibiotic susceptibility test was performed using the Gram-negative identification card of the Vitek 2 system (bioMérieux Inc., France), as well as an analysis of resistance genes, the effects of treatment with a light-emitting diode (LED) array using Radachlorin (RADA-PHARMA Co., Ltd., Russia), and transmission and scanning electron microscopy to confirm the biofilm-inhibitory effect of PDT.
Results
The antibiotic susceptibility test revealed multiple resistance to the antibiotics imipenem and meropenem in the carbapenem class. A class-D–type β-lactamase was found, and OXA-23 and OXA-51 were found in 100% of 15 A. baumannii strains. After PDT using Radachlorin, morphological observations revealed an abnormal structure due to the loss of the cell membrane and extensive morphological changes, including low intracellular visibility and small vacuoles attached to the cell membrane.
Conclusion
PDT involving a combination of LED and Radachlorin significantly eliminated the biofilm of multidrug-resistant A. baumannii. Observations made using electron microscopy showed that PDT combining LED and Radachlorin was effective. Additional studies on the effective elimination of biofilms containing multidrug-resistant bacteria are necessary, and we hope that a treatment method superior to sterilization with antibiotics will be developed in the future.
Introduction
Photodynamic therapy (PDT) is a cancer treatment method where photosensitizers responsive to laser light are administered to a tumor, and light of a specific wavelength is used to activate the photosensitizer to specifically damage cancer tissues using photochemical and photobiological mechanisms [1]. Photosensitizer molecules are activated by light of a specific wavelength and react with oxygen molecules in the body to generate reactive oxygen species [2,3]. Radachlorin (RADA-PHARMA Co., Ltd., Moscow, Russia), a promising photosensitizer that has been recently developed [4], is made from chlorophyll extracted from green algae that is activated by light of a 662-nm wavelength, exhibiting cytotoxicity and a penetrating power of approximately 10 mm. In addition, unlike Photofrin (QLT, Vancouver, Canada), a hematoporphyrin derivative, Radachlorin has the advantage of not exerting side effects on the human body, as it is made from chlorophyll [5].
Acinetobacter spp. infections represent a serious problem worldwide, accounting for 10% to 30% of all nosocomial infections in hospitals [6]. Currently, A. baumannii, the cause of more nosocomial infections, shows resistance to a wide range of antibiotics, including second- and third-generation cephalosporin agents, antipseudomonal penicillin agents, fluoroquinolones, and aminoglycosides, except for carbapenems [7–10]. A study examining the biofilm-forming ability of multidrug-resistant A. baumannii strains recently isolated from a clinical sample indicated that all resistant bacteria exhibited higher biofilm-forming ability than the standard A. baumannii strain ATCC 19606 [11]. In developed countries, over 60% of bacterial infections requiring treatment involve biofilm formation [12,13]. Bacteria with high biofilm-forming ability attach to host cells more easily or colonize medical devices and hospital environments, promoting horizontal gene transfer between bacteria in biofilms [14,15].
The most critical mechanism of carbapenem resistance acquisition in Gram-negative bacteria, including A. baumannii, is the inactivation of antibiotics through the generation of enzymes belonging to classes A, B, and D [16,17]. OXA-23–producing A. baumannii have been widely reported, primarily in Eurasia. There have been reports of carbapenem resistance caused by the overexpression of ISAba1 upstream of chromosomal OXA-51, and in European countries centered on the Mediterranean coast, there have been reports of OXA-48–producing enterobacteria [18]. The insertion sequence ISAba1 found in A. baumannii exists upstream of blaOXA-23 for generating OXA-23; it acts as a promoter and induces multidrug resistance by overexpressing blaOXA-23 [19]. However, reports on the effect of PDT on A. baumannii are scarce [20]. The aims of the present study were to identify the antibiotic susceptibility results and genetic pattern of A. baumannii, a multidrug-resistant bacterium that can be clinically isolated and is problematic in hospital-acquired infections, to determine whether PDT combining light-emitting diode (LED) and Radachlorin effectively eliminates its biofilm, and to analyze its efficacy through electron microscopy.
Materials and Methods
Experimental Strains
Fifteen multidrug- and carbapenem-resistant A. baumannii strains clinically isolated from December 1st to 28th, 2020 at a university hospital in Chungcheongnam-do Province were used. The strains were identified using the Gram-negative identification card of the Vitek 2 system (bioMérieux Inc., Marcy-l'Étoile, France).
Photosensitizer and Light Source
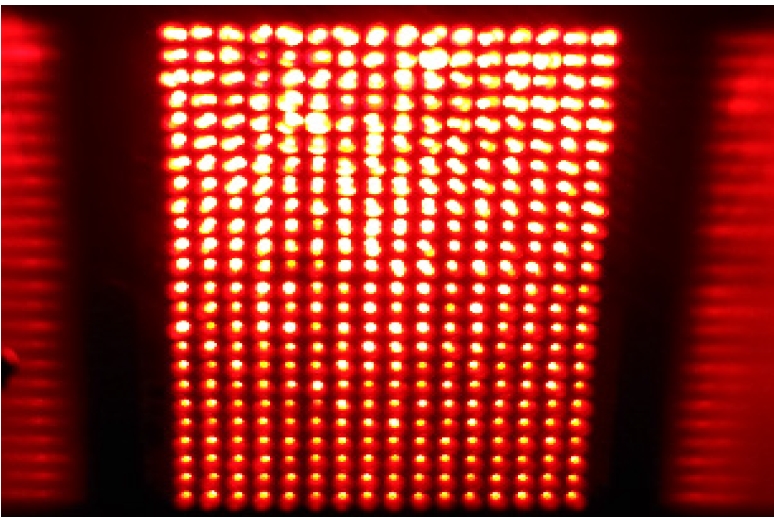
The photosensitizer Radachlorin (RADA-PHARMA Co., Ltd.) was used after diluting a liquid ampoule in Dulbecco’s phosphate-buffered saline (PBS; Hyclone, Logan, UT, USA) to a concentration of 1,000 μg/mL. Because the maximum absorption wavelength of Radachlorin is 662±3 nm, a LED array with an appropriate wavelength was self-produced. The LED used to irradiate the test tube had a cylindrical shape of 5Φ, and 384 (16×24) LEDs were installed with dimensions of 100 mm (width)×150 mm (length) (Figure 1).
Antimicrobial Susceptibility Tests
For strain identification, the test bacteria were subcultured on MacConkey agar at 37°C for 24 hours. The turbidity of the cultured colonies was adjusted to 0.5 McFarland standard using a loop [21]. The culture was then dispensed on the Gram Negative Identification card, and antibiotic discs of the Vitek 2 system impregnated with amikacin, ampicillin, aztreonam, cefazolin, cefepime, ceftazidime, ceftriaxone, ciprofloxacin, ertapenem, gentamicin, imipenem, levofloxacin, nitrofurantoin, piperacillin, and trimethoprim were used to test antibiotic susceptibility.
DNA Extraction and Analysis of Oxacillinase Class-D–Type OXA-23 and OXA-51
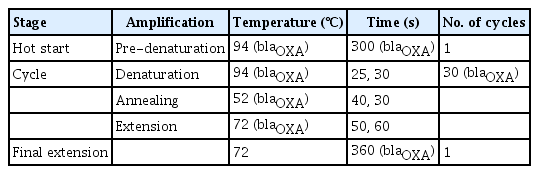
DNA was extracted using an EZ/Pure Cell/Tissue Mini kit (Enzynomics, Daejeon, Korea) and quantified using a Biodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA); when a purity of 1.0 or higher was quantitatively verified, the DNA sample was used for polymerase chain reaction (PCR) under the appropriate PCR conditions [21]. For base sequence conditions, blaOXA-23 and blaOXA-51 were verified to detect class-D–type β-lactamase. PCR was performed by adding 2 μL of template DNA and 1 U of Top DNA polymerase to 2.5 mM AccuPower PCR PreMix (Bioneer, Daejeon, Korea) containing lyophilized dNTPs (dATP, dGTP, dTTP, and dCTP) with a final concentration of 200 μmol. One microliter of each of the following primers was added (Table 1): OXA-23F (5′-GAT CGG ATT GGA GAA CCA GA-3′) and OXA-23R (5′-ATT TCT GAC CGC ATT TCC AT-3′) and OXA-51F (5′-TAA TGC TTT GAT) CGG CCT TG-3′) and OXA-51R (5′-TGG ATT GCA CTT CAT CTT GG-3′) [22]. The final reaction volume was adjusted to 20 μL with sterile distilled water. DNA was amplified using GeneAMP PCR system 9700 (Applied Biosystems, Waltham, MA, USA).
Biofilm Formation
Multidrug- and carbapenem-resistant A. baumannii was inoculated into MacConkey medium, streaked thrice using a loop, and cultured overnight in a 5% CO2 incubator (VS-9108MS; Vision Scientific Co., Ltd., Daejeon, Korea) at 37℃. After assessing the colonies for contamination, if no contamination was found, tryptic soy broth (Difco; BD Biosciences, San Jose, CA, USA) was used to make a strain suspension fluid in a polyethylene cap tube (SPL Life Sciences, Pocheon, Korea), and its turbidity was adjusted to 2.0 McFarland standard using a turbidimeter (DensiCHEK Plus; bioMérieux Inc.), resulting in a final concentration of 2.0×108 cells/mL. Furthermore, 100 μL of the strain suspension fluid was added to each well of a 96-well plate (Microtest 96 polyethylene; BD Biosciences) and incubated for 24 hours at 37℃. Next, the supernatant of the 96-well plate was discarded, and the biofilm spread on the bottom was washed thrice with PBS and dried naturally.
PDT for the Biofilm
After dividing into 6 groups (control, L+R−, L−R+100, L+R+1.0, L+R+10, and L+R+100) to perform PDT, 1,000 μg/mL of Radachlorin was collected from the freezer and thawed at room temperature for 15 minutes in a dark room while blocking light as much as possible. The control group was not treated using PDT as it did not form a biofilm. The L+R− group received LED treatment only. One Falcon tube was used for the L−R+100 group, in which the photosensitizer was directly diluted to a concentration of 100 μg/mL, and 100 μg was dispensed into each well, after which 900 μL of tryptic soy broth was dispensed into 3 Falcon tubes each, and the final volume of the diluted solution in the tube was adjusted to 1,000 μL. The photosensitizer was serially diluted 10-fold to final concentrations of 100, 10, and 1.0 μg/mL, dispensed into each well, wrapped in silver foil, and reacted for 1 hour in an incubator (VS-9108MS) at 37℃, following which PDT was performed for 1 h using a 630-nm LED. After PDT, the samples were washed thrice with PBS and naturally dried. Finally, 100 μL of safranin O was dispensed into each well, followed by a 30-s rest for staining, and the absorbance at 490 nm was measured using an enzyme-linked immunosorbent assay (ELISA) reader (Bio-Rad, Hercules, CA, USA).
Transmission Electron Microscopy Analysis
After PDT, a control strain-dilution solution and a strain-dilution solution with a photosensitizer concentration that inhibited biofilm formation were taken from their respective tubes and fixed with an equal amount of 2.5% glutaraldehyde (pH 7.4) at 4℃ for 12 hours. After centrifugation at 5,000×g for 20 minutes and discarding the supernatant, 2 mL of OsO4 solution was added to each obtained pellet and fixed for 1.5 hours. The pellets were then washed thrice with Dulbecco’s PBS without Mg2+ and Ca2+ (Cambrex, East Rutherford, NJ, USA) solution, dehydrated with ethanol, embedded in Epok812, and thermally polymerized at 35℃ for 10 hours at 45℃ for 10 hours, and at 60℃ for 48 hours. After preparing 500-nm semithin sections using an ultramicrotome, the block was stained using toluidine blue. Upon accurately identifying the area using a light microscope, the block was cut into a 70-nm ultrathin section. Finally, the morphology of the bacteria was observed using a transmission electron microscope (Hitachi 7700; Hitachi, Tokyo, Japan).
Scanning Electron Microscopy Analysis
After culturing A. baumannii in MacConkey medium for 24 hours, tryptic soy broth was added to a polyethylene cap tube (SPL Life Sciences) and adjusted to 2.0 McFarland standard using a turbidimeter (DensiCHEK Plus). Then, a strain suspension solution was made, and 500 μL of it was dispensed in each well of a 24-well plate (Microtest 24 polyethylene; BD Biosciences) and cultured for 24 hours at 37℃. The supernatant of the 24-well plate was discarded, and the biofilm spread at the bottom was washed thrice with PBS and dried naturally. Then, the photosensitizer at concentrations of 100, 10, 1.0 μg/mL was dispensed into each well, wrapped in silver foil, and reacted at 37℃ in an incubator (VS-9108MS) for 1 hour, following which PDT was performed for 1 hour using a 630-nm LED. The samples were then washed thrice with PBS and dried naturally. Subsequently, the biofilm-formation ability after PDT was compared with the control group and observed using a field-emission scanning electron microscope (FE-SEM, Hitachi S-4200; Hitachi). As pretreatment for FE-SEM analysis, the 24-well plates were treated for 30 minutes with a fixing agent comprising a mixture of 0.5% glutaraldehyde and 0.5% paraformaldehyde. After A. baumannii was fixed, it was washed with 0.1 M PBS, fixed with 1% OsO4 for 30 minutes, and rewashed with 0.1 M PBS. Then, conductive staining was performed for 30 minutes using 2% tannic acid, followed by washing with 0.1 M PBS; fixing with 1% OsO4 for 30 minutes; washing with 0.1 M PBS; dehydrating with 50%, 70%, 80%, 95%, and absolute ethanol for 10 minutes each; penetrating with tert-butyl alcohol for 30 minutes; and freeze-drying using a freeze drier (ES 2030; Hitachi). Finally, the freeze-dried sample was attached to a sample plate, coated with Pt–Pd alloy using an ion sputter (E-1030; Hitachi), and observed.
Statistical Analysis
One-way analysis of variance was performed to compare the observations of inhibition of biofilm formation between the control and experimental groups. When a significant difference was observed, the Tukey multiple range test and Bonferroni post hoc test were performed. PASW ver. 18.0 (SPSS Inc., Chicago, IL, USA) was used for the statistical analyses, and the significance level (α) was set to 0.05 for comparisons of the control group with the experimental group.
Results
Antibiotic Susceptibility Test
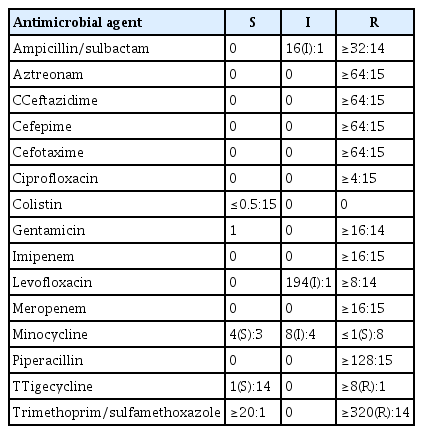
The antibiotic susceptibility results indicated that the bacteria showed multidrug resistance to imipenem and meropenem (both carbapenem antibiotics), 100% resistance to aztreonam (a monobactam antibiotic), 100% resistance to piperacillin (a third-generation penicillin antibiotic), and 100% sensitivity to tigecycline and colistin. Regarding cephalosporin antibiotics, 100% resistance was observed to the third-generation agents cefotaxime and ceftazidime. Furthermore,; 14 cases of resistance and 1 case of sensitivity to gentamicin; 14 cases of resistance and 1 case of moderate resistance to gentamicin; 14 cases of resistance and 1 case of moderate resistance to ampicillin/sulbactam; 14 cases of resistance and 1 case of moderate resistance to trimethoprim/sulfamethoxazole; 8 cases of resistance, 4 cases of moderate resistance, and 3 cases of sensitivity to the tetracycline antibiotic minocycline; 100% resistance to the fourth-generation cephalosporin antibiotic cefepime; and 14 cases of resistance and 1 case of moderate resistance to the new quinolone antibiotic levofloxacin were observed (Table 2).
Resistance Genotyping Analysis Results of A. baumannii
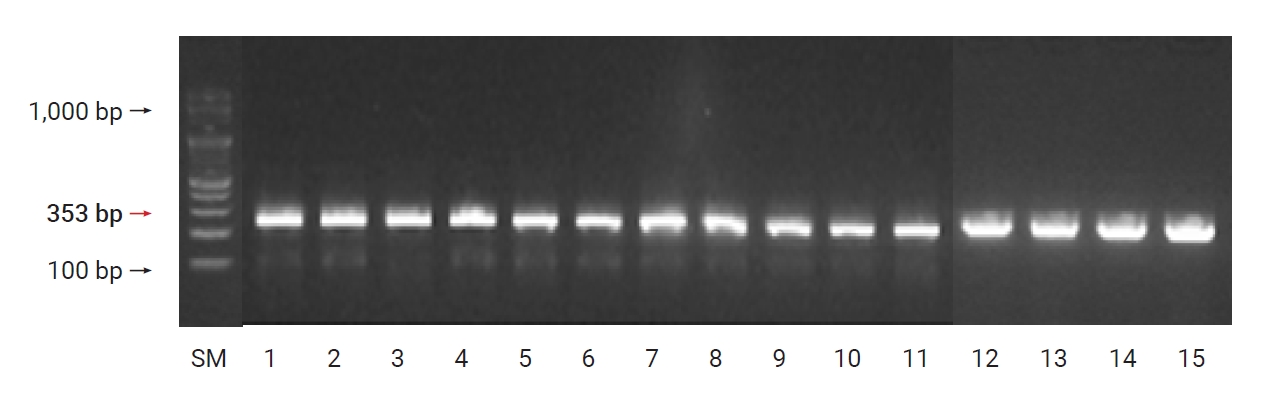
A. baumannii carbapenemase class-D–type β-lactamase was found, and OXA-23 and OXA-51 were found in all the 15 strains (100%) of A. baumannii (Figures 2 and 3).
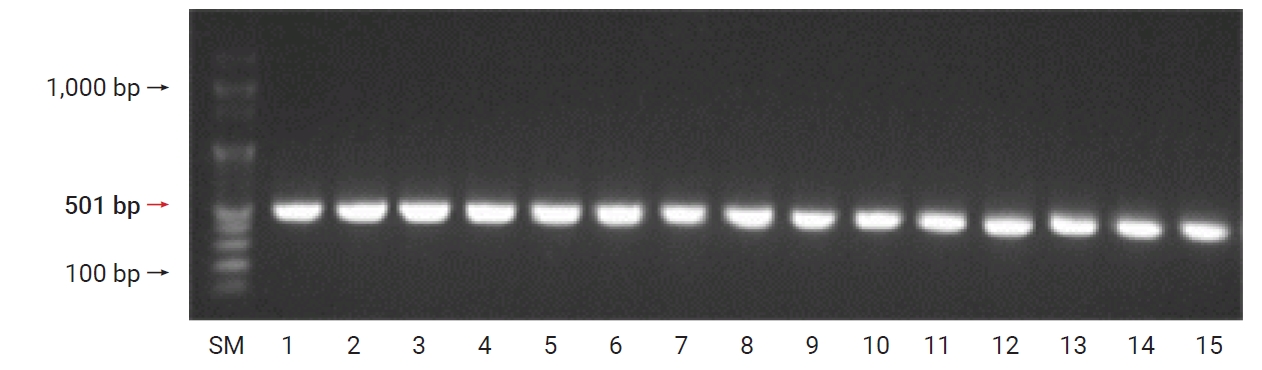
Detection of blaOXA-23 genes in Acinetobacter baumannii clinical isolates. Column 1 (size marker [SM]): 100 bp Plus DNA ladder; columns 2–16: different isolates containing the blaOXA-23 gene (501 bp).
Biofilm Formation Inhibition Test Results
Fifteen strains were incubated at 37℃ for 24 hours at a turbidity of 2.0 McFarland standard, and their biofilm-forming ability were observed in a 96-well plate after incubation. Eight strains (S1, S3, S5, S6, S9, S11, S13, and S15) formed biofilms, as observed with the naked eye, and the remaining 7 strains (S2, S4, S7, S8, S10, S12, and S14) did not form biofilms visible to the naked eye.
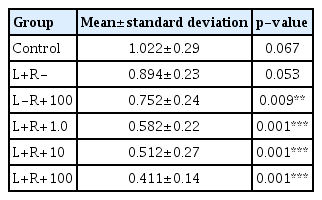
The average absorbance values of the biofilm observed with the naked eye measured using an ELISA (Bio-Rad) reader at 490 nm after PDT were as follows: control, 1.022±0.29; L+R–, 0.894±0.23; L−R+100, 0.752±0.24; L+R+1.0, 0.582±0.22; L+R+10, 0.512±0.27; and L+R+100, 0.411±0.14 (Table 3).
Cell Observation by Transmission Electron Microscopy
An A. baumannii strain was inoculated on 0.3% brain heart infusion nutrient agar, still-cultured at 37℃ for 48 hours, and the cultured bacteria were attached to a Cu-coated grid using 1% uranyl acetate (EMS Corp., Hatfield, PA, USA) solution and stained for 30 seconds. After washing with distilled water, the bacteria were observed using a transmission electron microscope (Hitachi 7700). The cell wall and cytoplasmic membrane of A. baumannii in the control group were clearly observed, and structures inside the cytoplasm also appeared without any alteration. However, the morphology of A. baumannii after PDT using a photosensitizer indicated at least partial loss of the cell membrane and abnormalities in its structure. Additionally, the extensive morphological changes after PDT included low intracellular visibility and small vacuoles attached to the cell membrane (Figure 4).
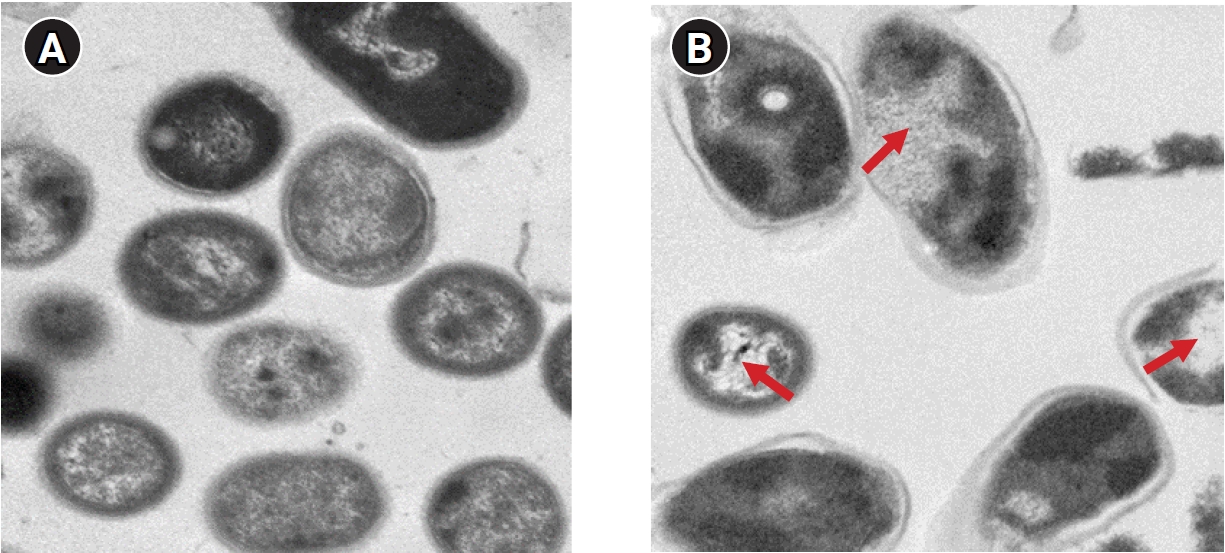
Ultrastructural changes in Acinetobacter baumannii treated with photodynamic therapy (PDT) by transmission electron microscopy. The untreated control group is shown in (A) (×5,000, 1 μm), and the PDT group in (B) (L+R+100; ×5,000, 1μm) was treated (red arrows) with a concentration of 100 μg/mL with Radachlorin following 630-μm light-emitting diode treatment for 1 hour.
Observation of Biofilm Formation Inhibition by Scanning Electron Microscopy
After the pretreatment of the 24-well plates that formed biofilms, the dried samples were attached to a sample plate, and an ion sputter (E-1030) was used to coat with them with a Pt–Pd alloy for observation under FE-SEM (S-4200). The biofilm had covered the entire control well, whereas wells that were subjected to PDT with Radachlorin and LED showed partial biofilm formation, especially in L+R+10 and L+R+100, followed by L+R+1.0, thus validating the efficacy of PDT (Figure 5).
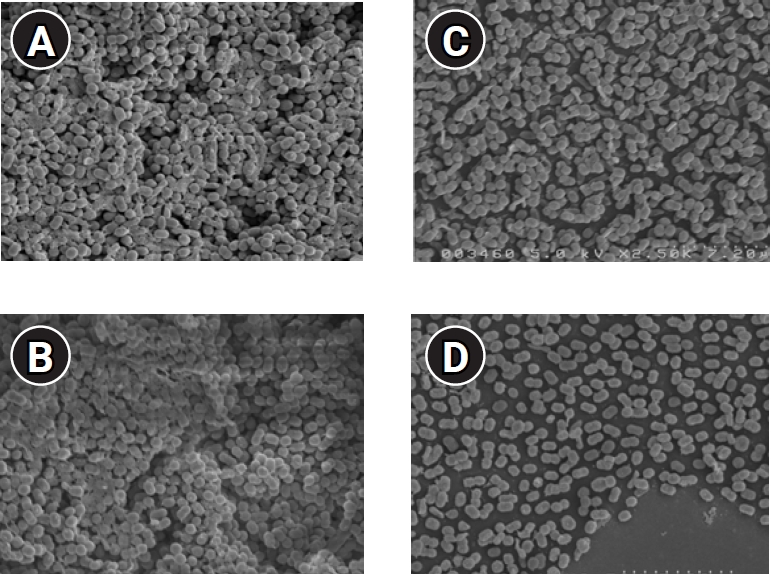
Field-emission scanning electron microscopy (FE-SEM) images of Acinetobacter baumannii biofilms established on a 24-well plate following 24 hours of incubation at 37°C in tryptic soy broth. (A) Control, (B) photodynamic therapy concentration group (L+R+1.0), (C) L+R+10, (D) L+R+100, FE-SEM pictures were taken at ×2,500 magnification; the inset white bar is representative of 7.2 microns.
Discussion
Pathogenic microorganisms, including A. baumannii, form biofilms in natural environments and medical insertion devices, causing problems such as widespread nosocomial infections of pathogenic bacteria [23]. These bacterial biofilms provide an environment for bacteria where they can survive for long periods from antibiotics and increase their pathogenicity [24]. The emergence and dissemination of carbapenem-resistant strains is considered a major clinical threat, as the imipenem resistance of an A. baumannii strain isolated in 1998 at a university hospital in Seoul was only 5% [25], whereas the ratio of imipenem-resistant A. baumannii increased from 4.3% in 2006 to 83.8% in 2010 [26]. Furthermore, A. baumannii isolated at Kosin University Gospel Hospital attained an imipenem resistance of 26.9%, suggesting the rapid spread of imipenem-resistant A. baumannii [27].
In a previous study, colistin resistance was reported to be present in 2.6% of 464 strains of A. baumannii; however, in the present study, 100% of A. baumannii exhibited imipenem resistance and no resistance to colistin [28].
Class D β-lactamases, which hydrolyze OXA-type carbapenem, including subtypes OXA-23, OXA-24/40, OXA-58, and OXA-51, are commonly isolated from A. baumannii [29]. Infection with OXA-23–producing A. baumannii has been reported in many regions worldwide, and OXA-23 is the most important mechanism by which A. baumannii acquires carbapenem resistance [18]. In this study, OXA-23 and OXA-51 were detected in 100% of the strains, suggesting their widespread presence. This indicates the necessity for ongoing, national-scale investigations for the genetic surveillance of antibiotic-resistant bacteria. The process of bacterial biofilm formation is partially regulated using quorum sensing, a cell-density signaling system, with a significant change of over 50% in the gene expression pattern during this process, and epithelial cell adhesion and colony-forming factors have been observed with multidrug-resistant A. baumannii [30].
In PDT, lethal bacterial damage is caused by 2 processes, the importance of which has been brought to attention. The first process is DNA damage, and the second is cytoplasm damage due to the weakening of the cell membrane transport system and inactivation of enzymes. One study showed the effect of PDT on Gram-positive and Gram-negative bacteria using a cyanide photosensitizer [31]. In another study, cell wall changes of the Vibrio vulnificus strain after PDT using toluidine blue were observed by transmission electron microscopy. The structure of V. vulnificus before PDT exhibited 1 long flagellum and clear hair-like cilia, but after treatment, several bubbles had formed in the cell wall [31]. In the present study, statistically significant results (p<0.001) were observed in the L+R+1.0, L+R+10, and L+R+100 treatment groups, in which the inhibition of biofilm formation was observed with the naked eye. When PDT was performed using both Radachlorin and LED, biofilm formation was inhibited significantly, verifying the therapeutic effect of PDT on multidrug-resistant bacteria.
Transmission electron microscopy indicated that several intracellular organs were damaged after PDT. It is believed that greater cellular damage results in greater reactive oxygen species generation. Damage to intracellular organs may have been caused by Radachlorin penetration into the cell and effective cell death by LED. Scanning electron microscopy exhibited significant biofilm reduction after PDT. Thus, while PDT against Gram-negative multidrug-resistant A. baumannii does not induce complete bacterial death, it effectively eliminates biofilm, and its statistically significant values suggest its therapeutic effect and reflect the superior therapeutic effect of PDT with LED and photosensitizers. Although the use of high concentrations of Radachlorin yielded superior results, a limitation is that the mechanism of the quorum-sensing signal transduction system was not studied; therefore, further research is required.
Although some studies have performed PDT in Gram-negative bacteria, few studies have examined its effect on multidrug-resistant bacteria. This study demonstrated the effectiveness of PDT on biofilm-producing multidrug-resistant bacteria. These findings suggest the possibility of using PDT as an alternative to antibiotics to inhibit bacterial growth, if more diverse studies are conducted on the removal of biofilms using multidrug-resistant bacteria, which pose a threat to medical institutions.
Conclusion
As a result of conducting PDT combining LED and Radachlorin, the biofilm of multidrug-resistant A. baumannii was eliminated, with significant results, and electron microscopy showed that the biofilm was more effectively eliminated in the L+R+10 and L+R+100 groups than in the L+R+1.0 group, proving that PDT combining LED and Radachlorin was effective. Although studies on PDT using multidrug-resistant bacteria are scarce, additional studies on the effective elimination of biofilm are necessary.
Notes
Ethics Approval
Not applicable.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Funding
This work was supported by the Konyang University Research Fund in 2020 (grant no. 20A0088).
Availability of Data
All data generated or analyzed during this study are included in this published article. Other data may be requested from the corresponding author.
Authors’ Contributions
Conceptualization: YBY; Data curation: CWS; Formal analysis: CWS; Funding acquisition: YBY; Investigation: YKK, PSK; Methodology: CWS, JLA; Project administration: JSK, YKK; Resources: YKK, YBY; Validation: CWS, YBY; Visualization: YBY; Writing–original draft: all authors; Writing–review & editing: all authors.