COVID-19 prediction models: a systematic literature review
Article information
Abstract
As the world grapples with the problem of the coronavirus disease 2019 (COVID-19) pandemic and its devastating effects, scientific groups are working towards solutions to mitigate the effects of the virus. This paper aimed to collate information on COVID-19 prediction models. A systematic literature review is reported, based on a manual search of 1,196 papers published from January to December 2020. Various databases such as Google Scholar, Web of Science, and Scopus were searched. The search strategy was formulated and refined in terms of subject keywords, geographical purview, and time period according to a predefined protocol. Visualizations were created to present the data trends according to different parameters. The results of this systematic literature review show that the study findings are critically relevant for both healthcare managers and prediction model developers. Healthcare managers can choose the best prediction model output for their organization or process management. Meanwhile, prediction model developers and managers can identify the lacunae in their models and improve their data-driven approaches.
Introduction
Healthcare refers to the organized provision of medical care to people and communities. It constitutes the efforts made by qualified and licensed practitioners to preserve or achieve physical, mental, or emotional well-being. Healthcare and medical facilities are regarded as making a significant contribution to the promotion of individuals’ health and well-being. The healthcare industry is responsible for manufacturing and distributing the drugs and services needed to safeguard, cure and sustain well-being. Providing healthcare for patients affected by coronavirus disease 2019 (COVID-19) has been challenging, especially in India and in Karnataka in particular. Several studies have been performed to understand the spread of COVID-19 and to deal efficiently with COVID-19 patients. The motivation of this study was to collate the available information on various prediction models and to choose accurate models for anticipating the number of cases. Many governments have collected and are trying to analyze data to be better equipped for providing healthcare to COVID-19 patients. The COVID-19 pandemic challenged healthcare facilities, with the sheer number of cases resulting in an acute shortage of capacity that constrained healthcare services [1]. A study was conducted to identify the best social media platform that can be employed for sentiment analysis and data mining, and the reported methods of data extraction and methodological consideration provide a basis for planning future studies [2]. State-of-the-art techniques for COVID-19 prediction algorithms are based on commonly used data mining and machine learning techniques to benefit the healthcare sector [3]. The management of the healthcare system focuses on the overall governance of public health services, including the appropriate and effective use of clinical infrastructure facilities, with a view to attaining the highest benefits for human health.
With the worldwide spread of the COVID-19 pandemic, which causes potentially severe respiratory illness, healthcare systems are facing challenges in order to provide appropriate treatment to support patients. In accordance with the goals of healthcare, there are several factors and aspects of the medical sector that must be actively planned and organized.
Adopting a multi-criteria decision framework, such as the Technique for Order of Preference by Similarity to Ideal Solution (TOPSIS) method, is an effective approach to prioritize COVID-19 patients that facilitates detection of the health conditions of asymptomatic carriers and helps stakeholders tackle the complex problem of COVID-19 [4–6]. The TOPSIS framework was developed based on machine learning and multiple-criteria decision-making via the subjective and objective decision by opinion score method to provide effective care and prevent the extremely rapid spread of COVID-19 from affecting patients and the medical sector [3].
Based on the findings of the systematic literature review (SLR), it is recommended that healthcare systems and stakeholders should use the best prediction model to forecast the number of cases and make the necessary arrangements for imposing social distancing and lock-down measures during the pandemic.
The present study provides insight into various prediction models and how to choose the best model in terms of maximizing accuracy and minimizing errors. This information will be vital in decision-making for government, the healthcare sector and other stakeholders. The findings of this study have implications for the quality of healthcare management. The health system is expected to perform well in all aspects of satisfying the needs of the customers whether those customers are patients, attending physicians, employers, or functional departments within an organization. The current study presents an SLR of papers published from January 2020 to December 2020. The study applied a specific set of inclusion and exclusion criteria to generate comprehensive tables reviewing the literature that contain information about various COVID-19 prediction models, the characteristics considered in prediction, sample size, and model accuracy.
Spread of COVID-19 (World-wide Scenario)
Pandemics are caused by pathogenic microorganisms (e.g., bacteria, viruses, parasites, and fungi) that tear through populations. The bubonic plague of the 14th century infected over 50 million people in Europe and the Spanish flu of 1918 infected a fifth of the world's population. Pandemic influenza, also termed H1N1 influenza/novel influenza/’swine flu,’ ravaged populations worldwide in more recent years [7].
COVID-19 is an infectious disease that affects the human respiratory system. In December 2019, the illness was first reported in Wuhan, the capital of China’s Hubei province. At the end of December 2019, a number of patients were admitted to hospitals with an initial pneumonia diagnostic test showing an unknown etiology. Since then, COVID-19 has spread around the globe. At the time of writing this paper (July 26, 2021), 90,698,044 cases of the virus had been recorded worldwide. COVID-19 was formally declared a global pandemic on March 11, 2020 by the World Health Organization (WHO). The top countries affected by COVID-19 are classified in terms of cases reported, deaths, and recovered cases (Table 1). The United States of America (USA), India, Brazil, Russia, France, United Kingdom, Turkey, Argentina, Colombia, and Spain are the top 10 countries affected by COVID-19. On January 13, 2020, the first case outside China was identified in Thailand [8,9]. The first case of COVID-19 was reported in the USA on January 23, 2020 [10].
Spread of COVID-19 in the Indian Context
India, which is the second most populated country after China, is the country in South Asia with the most COVID-19 cases. On January 30, 2020, India recorded the first case of the disease. Since then, cases have increased significantly and dramatically. In order to reduce the transmission of COVID-19, the government of India announced a nationwide lock-down starting on March 25, 2020, which continued for about 2 months. The number of COVID-19 cases as of July 31, 2021 has reached 197,548,856 confirmed cases and 4,213,071 cases. Within India, Karnataka is the second most strongly affected territory. In the early stages of the global pandemic, Karnataka registered fewer cases than most other Indian states. It was among the early states to deploy new equipment and tools as part of its infrastructure and containment initiatives. The first case in Karnataka was reported on March 9, 2020. The number of COVID-19 cases reported in Karnataka is 928,792 confirmed cases, 906,593 recovered cases and 12,142 deaths (as of January 11, 2021). The government of Karnataka incorporated a gradual lock-down, closing shops and offices, and shutting down inter-district and interstate journeys as part of the initiative to contain the outbreak. The period from March 24 to April 14, 2020 was phase 1 of the lock-down, with the strict restrictions on travel and social interaction. The second phase was from April 15 to May 3, and the third phase lasted from May 4 to May 17 [11]. Bengaluru, the capital of Karnataka, had more infections than other parts of the state. On March 9, 2020, the first COVID-19 case was identified in Bengaluru. As of January 11, 2021, the number of COVID-19 infections in Bengaluru amounted to 392,581 confirmed cases, 382,166 recovered cases, and 4,347 deaths. In terms of controlling the virus, Bengaluru has implemented various curfews, public awareness campaigns, and rigorous reverse-transcription polymerase chain reaction tests. The mapping of containment zones and predictive modeling conducted by Bruhat Bengaluru Mahanagara Palike (a local body) were vital factors for successfully controlling the pandemic (Figure 1).

Region-wise comparison of coronavirus disease 2019 (COVID-19) cases (as of January 2021), presenting the percentage share of COVID-19 cases reported as of January 2021. (A) World vs. India COVID-19 cases. (B) India vs. Karnataka COVID-19 cases. (C) Karnataka vs. Bengaluru COVID-19 cases.
COVID-19 is primarily transmitted by close contact with the droplets spread by sneezing, coughing, and talking to an infected person [12]. The initial stages in COVID-19 transmission have been attributed to human exposure in the wet animal market in Wuhan, where live animals are frequently sold, and it is speculated that this wet market was likely the main source of COVID-19 [13]. Efforts are being made to search for transitional carriers from which the infection might have spread to humans; however, regardless of the original source, COVID-19 has shown an unprecedented degree of horizontal spread. Person-to-person transmission takes place by close contact or through droplets spread by an infected person’s cough or sneeze [14].
WHO Definitions of Key Parameters
Confirmed case: A person with laboratory confirmation of COVID-19 infection, irrespective of clinical signs and symptoms.
Positive case (same as confirmed case): A person with laboratory confirmation of COVID-19.
Active cases: The value obtained by subtracting the number of recovered cases and the number of deaths from total number of positive cases.
Recovered cases: Those cured of COVID-19 and discharged from a healthcare facility, also referred to as “discharged.”
Death: For surveillance purposes, a COVID-19 death is characterized as a death resulting from a clinically compatible disease in a likely or confirmed case of COVID-19, unless there is a specific alternative cause of death that cannot be attributed to COVID-19 (e.g., trauma). There should be no time of full healing between sickness and death.
Symptoms: A moderate case is defined a confirmed case with fever, respiratory symptoms and radiographic evidence of pneumonia, whereas a case involving dyspnea or respiratory failure is defined as a severe case
Objectives
Owing to the wide spread of COVID-19 and its devastating effects on humans, several research groups have investigated various aspects of the virus, such as its epidemiological characteristics, socio-economic effects, and factors and parameters aiding the spread of the virus. The present work is an SLR with the following objectives: (1) To systematically review the prediction models that have been developed for COVID-19; (2) To analyze the various COVID-19 prediction models that are currently available; (3) To synthesize and extract useful results and conclusions about the COVID-19 prediction models.
Methods
An SLR is a supplementary methodology used to help evaluate studies by capturing principal analyses on the basis of specific criteria. An SLR is carried out on the basis of previous similar studies through a systematic review. The purpose of an SLR is to summarize the studies carried out and to identify gaps between previous studies and current studies.
Okoli [15] stated that an SLR is “a systematic, explicit, detailed and repeatable approach to identify, assess and analyze the existing body of work by researchers, scholars and practitioners.” According to Tranfield et al. [16], an SLR is considered as a “fundamental scientific activity.” Moher et al. [17] presented a checklist for Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA). The objective of this SLR was to understand further the mechanisms and analyses used in prediction models for COVID-19 infections. The research time period for this study was from December 2020 to January 2021. This study was conducted in 4 phases: (1) the development of literature search strategies, (2) the formulation of inclusion and exclusion criteria, (3) quality assessment, and (4) analysis and conclusion.
Research Questions
The wide spread of the COVID-19 pandemic has resulted in illness and loss of life on a global scale. Research teams have worked on various models to understand the spread of the virus and make data-driven predictions. For the purpose of this SLR, we articulated a research question (RQ) to help focus on the main issue. The motivation and RQs of this study were as follows:
Motivation: To identify methods, techniques, models that support the prediction of COVID-19 infections.
RQ1: What factors support the prediction of COVID-19 infections?
RQ2: What methods and techniques are followed in data-driven modeling for predicting COVID-19 infections?
Inclusion and Exclusion Criteria
Current search engines provide a high level of recall, which leads to a large number of irrelevant resources being retrieved. Therefore, for effective results, a researcher must follow a systematic search strategy. This stage of an SLR screens the literature to find the relevant literature on the basis of particular criteria. In this study, 3 inclusion and exclusion criteria for identifying relevant content and restricting irrelevant content were adopted. The first inclusion criterion was the type of document: only published documents were included, whereas manuscripts under review and unpublished manuscripts were excluded. The domain (i.e., the subject area identified for the study) was the second screening criterion; the authors included documents containing prediction models developed for or used in the COVID-19 domain, while other documents were excluded. The last screening criterion was the language in which the document was released. In order to avoid confusion and complexity related to translation, only documents available in English were included, while documents in other languages were excluded (Table 2).
Databases and Search Strategies
The terms were searched in several databases (Google Scholar, Scopus, Publish or Perish, and Web of Science [WoS]). The search terms are as follows: prediction models, COVID-19, Coronavirus, SARS-CoV, SARS-CoV-2, healthcare, healthcare system, survival model, medical care. Various combinations of the search terms were used to retrieve resources in particular databases. Some of the search strings used are as follows—“Prediction models” AND “COVID-19”; “COVID-19 Datasets” AND “Prediction modeling”; “Predictive Analysis” AND “COVID-19 data” OR “Predictive Analysis” AND “Corona Virus”.
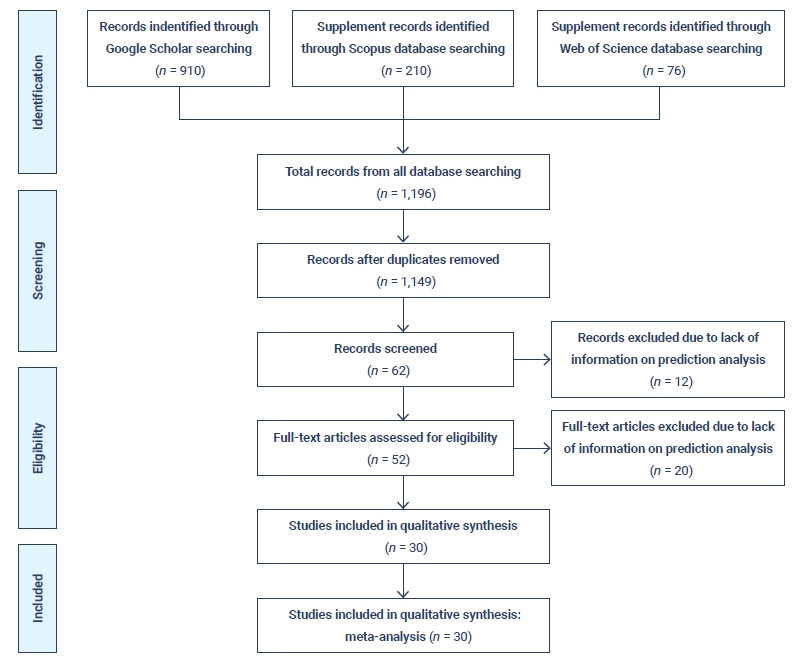
After applying the inclusion and exclusion criteria, 1,196 documents were retrieved, of which 47 were duplicates. Therefore, a total of 1,149 documents continued to the second stage of scrutiny and quality assessment (Table 3). The percentage shares of articles from various databases in the initial, screening, and acceptance stages of the document selection process are illustrated.
In the initial phase, out of the total number (i.e., 1,196 documents) of retrieved documents, Google Scholar accounted for 77%, Scopus contained 17%, and WoS had 6% (Figure 2A). After the initial screening, 62 documents were included for further consideration. During the screening phase, 52% of the initially included documents were retrieved from Google Scholar. Out of the remaining 30%, Scopus and WoS had an 18% share each (Figure 2B). Out of the total accepted documents, 70% were retrieved from Google Scholar, 14% from Scopus, and 16% from WoS (Figure 2C).

Database's percentage share of COVID-19 cases reported as of January 2021.
(A) Document selection (initial). (B) Document selection (screened). (C) Document selection (included). Document selection was carried out based on selection criteria.
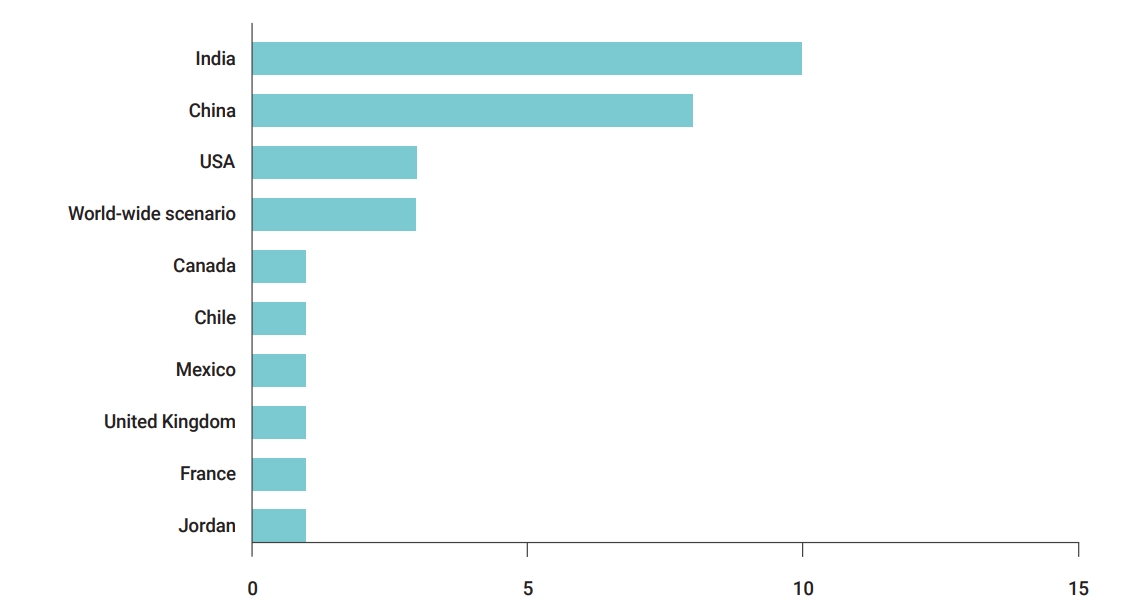
The present study focused on publications dealing with COVID-19 prediction models across the world. This review was conducted in January 2021. The country of a research/case was defined by the affiliations of authors in the paper, and a limited research level was observed for several countries (e.g., Canada, Chile, France, Jordan, etc.). Given our particular focus on the spread of the pandemic in India, the highest number of publications was from India and China (Figure 3).
Quality Assessment and Coding
Quality evaluation of a phenomenon is conducted as a systematic way to avoid biases and errors. Thereby, an SLR includes quality assessment as an essential step. In this study, in the initial phase, 1,196 documents were chosen. Based on their titles, these documents were further analyzed and 62 documents were screened. The content was scrutinized on the basis of the title, abstract, introduction, and conclusion and 30 studies were finally selected for the review.
Related Literature
Prediction Models
A prediction model is a method of becoming aware of a future scenario beforehand based on available data. Predictive modeling mainly uses statistics to predict outcomes [18]. Forecasting in the COVID-19 pandemic allows medical professionals to better manage facilities and to validate the use of medical and financial resources. It is essential to systematically assess the predictive outcomes of 1 or more prediction models in order to analyze the prediction accuracy of a framework across different study populations, ecosystems, and locations and to assess the need for further developments or improvements of a model [19]. In this paper, we present a systematic review and analysis of these models as presented in the literature.
Related Works
Coronaviruses are among the main pathogens that predominantly affect the human respiratory system. The focus of the literature review was, therefore, to outline the predominant variables and methodology used in studies related to the spread of the virus. People with prevalent illnesses such as diabetes, hypertension, diabetes, stroke, heart, or kidney failure, as well as elderly people with impaired immune systems, are at an increased risk of infection [20]. Closed areas with low ventilation and airflow may increase the risk of infection. The spread of the virus is believed to occur through respiratory droplets from coughing and sneezing, as with other respiratory viruses, including influenza virus and rhinoviruses. Aerosol transmission is also possible in case of protracted exposure to elevated aerosol concentrations in closed spaces [21].
Several reports have defined a series of variables in terms of quarantine facilities, laboratory testing facilities, and healthcare capability, contributing to state preparedness to fight the pandemic. The most important and successful of these factors must be explored as an urgent solution to the pandemic. The availability of open data sets corresponding to different variables helps to accelerate studies and forge cooperation [22]. Environmental factors, such as pollution and basic sanitation, were considered in some studies. Several studies have also taken into consideration deaths due to COVID-19 and other demographic information [23,24]. Other studies and theories have pointed to comorbidities as a key factor in the number of COVID-19 cases [25,26]. Without considering comorbidities, fatalities may be mistakenly interpreted as exclusively COVID-19 deaths. Researchers from many universities in the USA have successfully predicted COVID-19 deaths. One such study was conducted at Columbia University and the CDC (2020), in which “death” was used as an exponential function and a social distance parameter prediction was made using a susceptible-exposed-infectious-removed (SEIR) meta-population model.
Since the very beginning of the COVID-19 pandemic, numerous researchers have attempted to construct statistical models of the COVID-19 pandemic, as can be seen from a primary review of existing models. There are several differences in scope, assumptions, forecasts, the effects of interventions, and their impact on health services [27]. A PRISMA flow diagram based on the identification of studies from various databases, screening, and the eligibility and inclusion criteria is presented in Figure 4.
SLR on COVID-19
In the context of the COVID-19 pandemic, people across the world are using various methods to explore prediction models with the goal of addressing the problems caused by the pandemic. The motivation for this SLR was to help researchers across the world study the various prediction models that have been created by numerous authors from multiple countries by providing information on a comprehensive range of models in one place. A systematic review is a compilation of various studies related to a single topic. It aims to provide a comprehensive and unbiased review of all the relevant studies in a given field. Our SLR was conducted to determine which prediction models are currently available, and the objective of the study was to identify the various methods used to develop different types of prediction models and to conduct an effectiveness or quality assessment of the models, which helps to evaluate their accuracy. It is hoped that this SLR will help healthcare workers and researchers wisely and confidently choose accurate prediction models to facilitate healthcare management by arranging medical facilities and equipment. Researchers or scholars can enhance their research program by using this SLR to obtain up-to-date information on the various techniques used in prediction models, as well as their efficiency and accuracy. All currently available prediction models for COVID-19 were systematically reviewed and critically appraised. There are currently a number of diagnostic and prognostic models for COVID-19, all of which show moderate to excellent discrimination. To explore the different prediction models and find the best-suited model in terms of providing high accuracy while minimizing the burden on the healthcare system and improving care for patients, both the diagnosis and prognostic evaluation of diseases need to be improved. This study will influence decision-makers in various aspects.
The selected papers deal with different techniques used to build predictive models for the spread of COVID-19. Various techniques are used for the modeling and to present results. Quantitative assessments were also evaluated based on the papers’ presentation of the percentage success/accuracy rate or error rates in statistical and regression models. This SLR sums up the research work of different prediction model developers in detail. In this SLR of prediction models related to the COVID-19 pandemic, we identified 30 studies with various prediction models. Among the 30 papers, the most cited ones were found to be those authored by Chinese researchers, followed by papers authored by Indian researchers and then papers authored by USA-based researchers (Table 4) [12,28–56].
To identify the likelihood of future results based on historical data, predictive analytics uses data, statistical algorithms, and different techniques such as machine learning, autoregressive integrated moving average (ARIMA) models, SEIR models, and long short-term memory (LSTM) models. The present SLR also classified papers on the basis of the techniques used (Table 5) [12,28–56]. The most commonly used techniques used in predictive modeling and analysis were as follows:
Machine learning
Machine learning is a technique used in which computers evaluate a data set and learn from the insights they gather. An artificial neural network is simulated by the use of complex algorithms that allow machines to classify, interpret, and understand data, and then use the insights that have been obtained to solve problems or make predictions. Common examples of machine learning include classification models, forecasts, medical diagnosis, image processing, regression, chatbots, and recommendation engines. Machine learning is a different branch of programming and is known to be an emerging technology.
ARIMA models
ARIMA models can be built in an array of software tools, including Python. These models are used in statistics and econometrics to measure events that happen over a span of time. ARIMA models predict future data in a series using past data. An ARIMA model can be constructed for any number series that display patterns and is not a random event series. For example, sales data from a footwear store would be an example of time series data because the data are collected over a period of time. One of the key characteristics is that the data are collected at constant, regular intervals [57].
SEIR models
SEIR models are commonly used for assessing infection data during the different phases of an infectious outbreak. SEIR models are among the most widely adopted mathematical frameworks to describe disease dynamics and forecast potential contagion scenarios. After an infectious disease outbreak, a SEIR model can be helpful in determining the efficacy of different interventions, such as lock-downs. These models are based on a series of complex ordinary differential equations that take into account the number of people who are sick, the pattern of people who recover over time after sickness, and the people who die [58].
LSTM models
LSTM models are a type of recurrent neural network (RNN) used to predict new infection numbers over time by processing and forecasting several issues related to time series. With repeating modules like an RNN, an LSTM model has a chain-like structure, except that instead of a single neural network layer as in RNNs, an LSTM model has 4 layers that communicate in a slightly different manner, each of which performs its own special network role. In an LSTM cell, each repeating module has a cell state. Through using various gates in the cell, the LSTM cell has the power to add or subtract information to the cell state. There are 3 gates for the standard LSTM cell that control the sum of data input or output to/from the cell state and protect the cell state.
Regression models
Regression analysis is a method of quantitative research that is used in studies modeling and analyzing several variables, where a dependent variable and 1 or more independent variables are included in the relationship. In basic terms, regression analysis is a mathematical approach used to evaluate the existence of the relationship between a dependent variable and 1 or more independent variables [59]. The 2 most widely used regression analyses are: (a) Logistic regression: in logistic regression, an independent variable is used to estimate the dependent variable. (b) Support vector regression (SVR): SVR provides the flexibility to determine how much error is suitable in a model and to find an appropriate line (or hyperplane in higher dimensions) to match the results.
GLEM models
Global epidemic and mobility (GLEM) models are being used in a number of COVID-19 related studies and analyses. These models involve a stochastic computational framework that combines high-resolution demographic and mobility data across the globe to predict the epidemic distribution across the globe. The goal of the GLEM model is to optimize versatility in specifying the disease compartment model and configuring the simulation scenario. It allows the user to set a number of criteria, including compartment-specific features, transition values, and environmental effects [60].
Conclusion
This study identified the core literature on prediction models for COVID-19. The aim of this research was to review and analyze the articles in the literature related to prediction models for COVID-19. A prediction model is a method for predicting the future scenario based on present facts. This SLR was based on a manual search of 1,196 papers published from January to December 2020, out of which 30 documents were selected on the basis of inclusion and exclusion criteria. Our SLR was conducted to explore which prediction models are currently available, with the goals of identifying various methods used to develop different types of prediction models and to conduct an effectiveness or quality assessment of models, which helps in evaluating their accuracy.
Based on this review, it is critical for statistical methods to be extensively used to predict the spread of infection. The LSTM [35] approach was used to track COVID-19 cases and to help government officials and policymakers in preparedness, with a root mean square error (RMSE) of 45.72. An ARIMA [47] model was used to predict the spread of COVID-19 infection with an average RMSE 44.81, followed by machine learning, artificial intelligence, and hybrid models. Lastly, in a few of the studies, mathematical modeling and network-based forecasting were used. SEIR models are among the most widely adopted mathematical frameworks to describe disease dynamics and forecast potential contagion scenarios. This SLR provides detailed information about various COVID-19 prediction models that can be adopted by researchers. This information can be used by healthcare professionals and by local government bodies in order to make decisions for managing healthcare facilities accordingly.
Notes
Ethics Approval
Not applicable.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Funding
None.
Availability of Data
Data for literature review was taken from Google Scholar, Scopus, and Web of Science. All data generated or analysed during this study are included in this published article. For other data, these may be requested through the corresponding author.
Authors’ Contributions
Conception: all authors; Design: all authors; Supervision: RS, DRS; Literature review: SMS, NSK, PPM; Writing–original draft: all authors; Writing–review & editing: all authors.