Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 11(4); 2020 > Article
-
Original Article
Risk Assessment Program of Highly Pathogenic Avian Influenza with Deep Learning Algorithm - Hachung Yoona, Ah-Reum Jangb, Chungsik Junga, Hunseok Kob, Kwang-Nyeong Leea, Eunesub Leea
-
Osong Public Health and Research Perspectives 2020;11(4):239-244.
DOI: https://doi.org/10.24171/j.phrp.2020.11.4.13
Published online: August 31, 2020
aVeterinary Epidemiology Division, Animal and Plant Quarantine Agency, Gimcheon, Korea
bKorea Telecom, Seoul, Korea
- *Corresponding author: Hachung Yoon, Veterinary Epidemiology Division, Animal and Plant Quarantine Agency, 177 Hyeoksin 8-ro, Gimcheon, 39660, Korea, E-mail: heleney@korea.kr
• Received: March 22, 2020 • Revised: May 2, 2020 • Accepted: June 22, 2020
Copyright ©2020, Korea Centers for Disease Control and Prevention
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Abstract
-
Objectives
- This study presents the development and validation of a risk assessment program of highly pathogenic avian influenza (HPAI). This program was developed by the Korean government (Animal and Plant Quarantine Agency) and a private corporation (Korea Telecom, KT), using a national database (Korean animal health integrated system, KAHIS).
-
Methods
- Our risk assessment program was developed using the multilayer perceptron method using R Language. HPAI outbreaks on 544 poultry farms (307 with H5N6, and 237 with H5N8) that had available visit records of livestock-related vehicles amongst the 812 HPAI outbreaks that were confirmed between January 2014 and June 2017 were involved in this study.
-
Results
- After 140,000 iterations without drop-out, a model with 3 hidden layers and 10 nodes per layer, were selected. The activation function of the model was hyperbolic tangent. Precision and recall of the test gave F1 measures of 0.41, 0.68 and 0.51, respectively, at validation. The predicted risk values were higher for the “outbreak” (average ± SD, 0.20 ± 0.31) than “non-outbreak” (0.18 ± 0.30) farms (p < 0.001).
-
Conclusion
- The risk assessment model developed was employed during the epidemics of 2016/2017 (pilot version) and 2017/2018 (complementary version). This risk assessment model enhanced risk management activities by enabling preemptive control measures to prevent the spread of diseases.
- Keywords: avian influenza; deep learning; risk assessment
- Avian influenza viruses (AIV) belong to the family Orthomyxoviridae and the genus influenza virus A [1]. AIV is known to spread from flock-to-flock and farm-to-farm in a number of ways. Humans are involved in the mechanical transfer of the pathogens, with the movement of personnel and equipment including vehicles [2,3]. On the assumption that animals and materials are transported by vehicles and driven by people, vehicles can act as a fomite of virus transmission. Visit records of livestock-related vehicles can be used as a source of data to trace farm-to-farm transmission of viruses. Moreover, a combination of category and location information can reflect the epidemiological characteristics of vehicles. This would include the location of stops or parking places on the farm, the number of vehicle passengers (visitors), and the visitor’s reason for being there (i.e. contact with animals, hygiene information) [4]. Data on the movement of vehicles can be measured by global positioning systems and extracted through the “internet of things”, which can collect data directly from devices connected to the internet. Data analysis can produce descriptive and predictive outputs with a timely dynamic and complexity to support intelligent informed decisions [5].
- A risk assessment program was developed to estimate the risk of highly pathogenic avian influenza (HPAI) at the farm level, using the visit records of livestock-related vehicles in the Korean animal health integrated system (KAHIS), a national database. The program was devised by the Animal and Plant Quarantine Agency (APQA) of the Korean government and a private corporation (Korea Telecom, KT). The pilot program was implemented during the HPAI epidemic of November 2016 to April 2017, and after being adjusted, the amended program was implemented during the HPAI epidemic of November 2017 to March 2018. This risk assessment program is available in a frame of the graphic user interface of the KAHIS. In this study, the process of model development and validation using real outbreak data, and its use in the application to a HPAI epidemic in the Republic of Korea is described.
Introduction
- 1. KAHIS
- KAHIS [6] is a web-based database which integrates all data associated with livestock and animal health in the Republic of Korea. The records of livestock-related vehicles’ (hereafter referred to as vehicles) visits to livestock facilities are a part of the data collected by KAHIS. When a vehicle crosses the boundaries of a previously determined set of global positioning system coordinates, a record of information, such as time of visit, facility details (type, name, owner, location, and animal species when the facility is a farm), and vehicle details (i.e. registered number, category of use for the vehicle, driver, owner, car number, vehicle type) is generated. As of December 2019, 60,500 vehicles were registered with KAHIS [6]. The vehicle’s location was registered at the municipality level with 2 criteria: operators (passengers) and materials carried. The categories of material carried included live animals, raw milk, eggs, egg trays, veterinary pharmaceuticals, feed, forage, husk, livestock manure, compost, and byproducts of poultry. The categories for the operator included veterinarian, vaccine practitioner, artificial inseminator, consultant, machine repairman, laborer (for example for loading and unloading poultry animals), and facility operations manager (farmer). In the KAHIS, livestock facilities were classified into farm, abattoir, raw milk collection house, feed mill, live animal sales field, animal capability testing agency, hatchery, egg grading and packing center, and livestock manure treatment plant. Poultry farms were categorized as broiler duck, breeder duck, breeder chicken, layer hen, broiler chicken, Korean native chicken, quail, and others.
- 2. Study data
- The risk assessment program was developed with data from the visit records of the confirmed HPAI outbreaks on farms and any outbreak-related farms (hereafter referred to as related farms), that is, farms that have an epidemiological linkage with a farm, or farms confirmed to have an HPAI outbreak. HPAI outbreaks involved 544 poultry farms (307 farms with H5N6, and 237 with H5N8) that had available visit records amongst 812 HPAI outbreaks confirmed between January 2014 to June 2017. Data were extracted from the KAHIS and transferred to the APQA’s big data platform through an extract-transform-load process. Raw data were available in the KAHIS and meta-data were stored in the Veterinary Epidemiology Division of the APQA.
- Before developing the risk assessment program, characteristics of the visit record data were analyzed to identify any dangerous contacts. The attack rate of dangerous contacts was estimated as the number of HPAI “outbreak” farms amongst the poultry farms visited by each vehicle. A dangerous contact [7] was defined as a contact between 2 “outbreak” farms, made by a vehicle that visited both farms, where the visits satisfy the following 3 conditions: firstly, a vehicle visited the source farm within 21 days before the outbreak date of the source farm; secondly, the same vehicle visited the receiver farm, within 21 days before the outbreak date of the receiver farm; thirdly, the 2 visits were no more than 21 days apart. The period of 21 day was selected due to the assumed maximum incubation period, according to the standard operation procedure of HPAI in Korea [8]. The outbreak date refers to the date of collecting the specimen in which HPAI virus was confirmed.
- 3. Risk assessment program
- The risk for a HPAI outbreak was predicted for the related farms. The visit records of vehicles were traced from the date the first contact was made with the “outbreak” farm until the date of the outbreak at the farm. Risk was first recognized for each visit made by a vehicle to a farm at a specific time. Then a farm-level risk was regrouped day by day, for 22 days (21 days prior to the outbreak, plus the outbreak date) whilst taking the previous day’s risk into account. The final risk for a related farm was determined on the outbreak date of the “outbreak” farm.
- The risk assessment program was a type of deep neural network (DNN) model developed using the multilayer perceptron (MLP) method [9], and built with the R Language (https://www.r-project.org/). The response variable of the model was in binary form (outbreak/non-outbreak). The explanatory variables contained information on farms (animal species, number of heads, farming type, geographical coordinates, and history of HPAI outbreaks), environment (farm density), vehicles (purpose of operation, owner, and driver), and visit records (time of visit, time interval between the visits to the source and receiver farms, visits to any livestock facilities between the visits to the 2 farms, frequency of visits, interval between visit and outbreak dates) and associated livestock-related facilities. The explanatory variables were either categorical or continuous. The categorical explanatory variables were transformed into a dummy format to be considered in the risk evaluation model. Details of the explanatory variables are described in Supplementary Table 1. Predicted risk values were produced by the model as a continuous value between 0 and 1.
- The risk assessment program developed using the MLP method consisted of 3 datasets derived from the original dataset: training (50%), validation (30%) and test (20%). For the training step, the relationship between the visit characteristics of the vehicles and the HPAI outbreak on a farm was analyzed under a combination of candidate parameters including the number of hidden layers, number of nodes per hidden layer, and proportion of drop-out and type of activation function [10]. The optimal models were selected based on precision and recall of the test to give an F1 measure. The value of precision (positive predictive value in epidemiology) p, is the fraction of the outbreak over all outbreak predictions [(where tp is true positive and fp is false positive)]. Recall (sensitivity) r, was the fraction of predicted outbreak over all outbreaks calculated [(where fn is false negative)]. The F1 measure was the harmonic mean of precision and recall ( ) [11]. Models with the highest values of precision, recall, and F1 measure were considered for the final selection.
- To validate the performance of the risk assessment program, the predicted risk values were compared between “outbreaks” and “non-outbreaks” on farms using the Z-test. Recall, precision and F1 measure were estimated with 101 different cut off values from 0 to 1, with an interval of 0.01. A positive likelihood ratio (PLR+=tp/fp) was also calculated by 0.01 of the predicted risk values and posterior probability of an outbreak (PPO+) was estimated (PPO+=predicted risk values by model × PLR+) [12,13].
- 4. Ethics approval
- Ethical policies of the journal have been adhered to. No ethical approval was required for this study because the data used did not contain private information.
Materials and Methods
- 1. Dangerous contacts
- The visit records of vehicles going to poultry facilities registered between 26th December 2013 to 2nd April 2017 were examined. As this period corresponded to the 21 days prior to the date of the first outbreak (16th January 2014) and the date of the last outbreak (2nd April 2017), a total of 58,026 visit records were generated by 34,343 vehicles in association with 544 HPAI “outbreak” farms. There were 23,174 farm-to-farm links between source and receiver. There were 3,208 cases of dangerous contacts identified (5.5% of 58,026 visit records), in which 442 vehicles made contact between 338 sources and 357 receiver farms. The dangerous contacts were generated by 1,710 sets of source-vehicle-receiver combination. Most of the combination sets were the same type of farm (1,569 sets, 91.8%) and the same animal species (1,669 sets, 97.6%). The most related categories of vehicles carried feed, live animals, eggs, and husk in decreasing order, with the number of visits for both total and dangerous contacts. Meanwhile, the highest attack rates were observed for vehicles carrying eggs and livestock compost (Table 1).
- 2. Model selection and validation
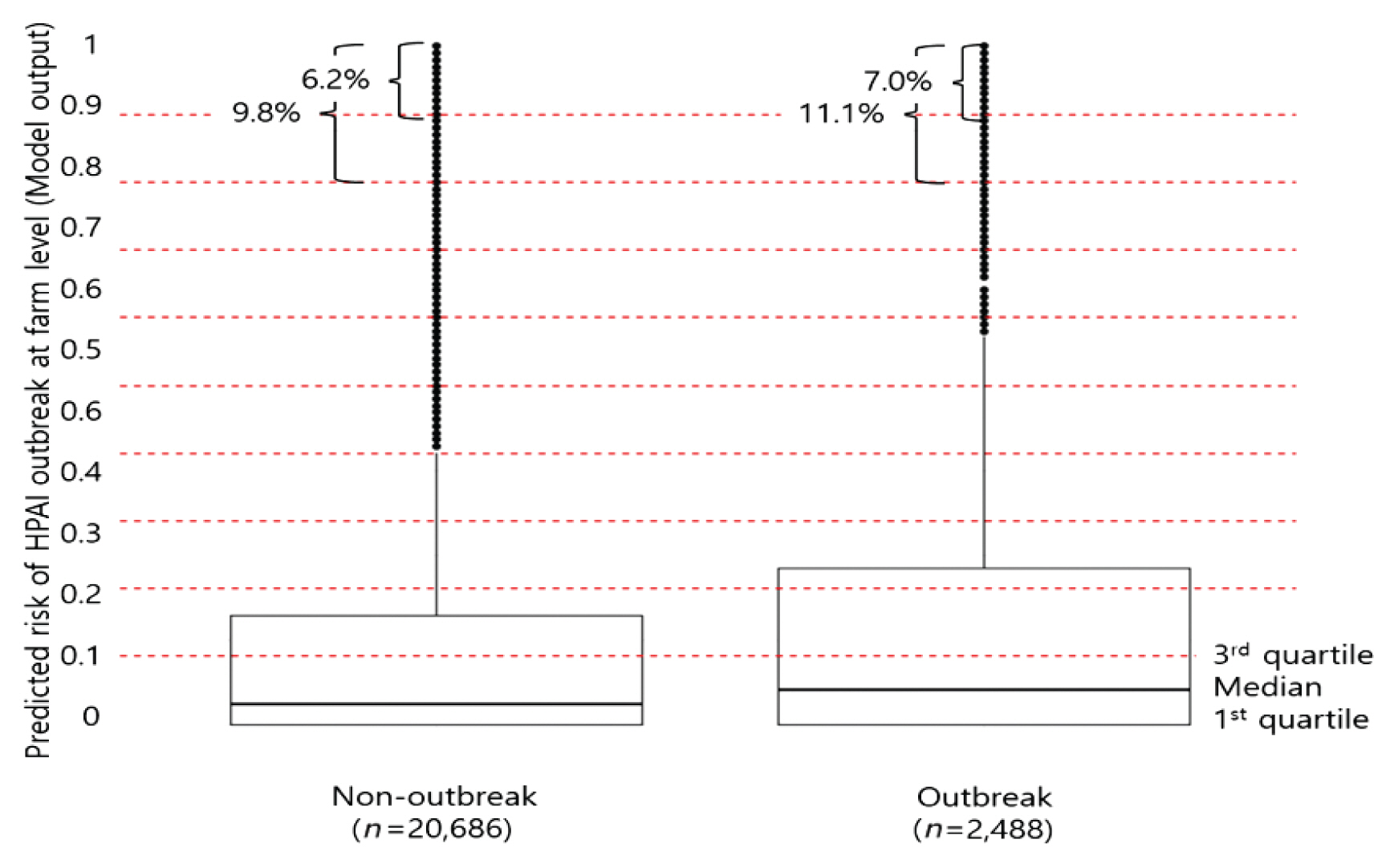
- After 140,000 iterations without drop-out, a model with 3 hidden layers and 10 nodes per layer was selected. The activation function of the model was hyperbolic tangent. Precision, recall, and F1 measures were 0.41, 0.68 and 0.51, respectively, at validation, and 0.54, 0.90 and 0.65, respectively, at the test step. Amongst 23,174 farm-to-farm links through visiting vehicles, 2,488 (10.7%) were going to “outbreak” farms and 20,686 (89.3%) were going to “non-outbreak” farms. The predicted mean risk values were higher for the outbreak (0.20 ± 0.31) than “non-outbreak” (0.18 ± 0.30) farms (p < 0.001). The median and third quartiles for the predicted risk values were 0.03 and 0.16 for “non-outbreak” farms, while they were 0.05 and 0.23 for the “outbreak” farms. The proportion of the predicted risk values of 0.8 and above, was 9.8% for the “non-outbreak” farms, while it was 11.2% for the “outbreak” farms, and risk values of 0.9 and above, were 6.2% and 7.0%, respectively (Figure 1).
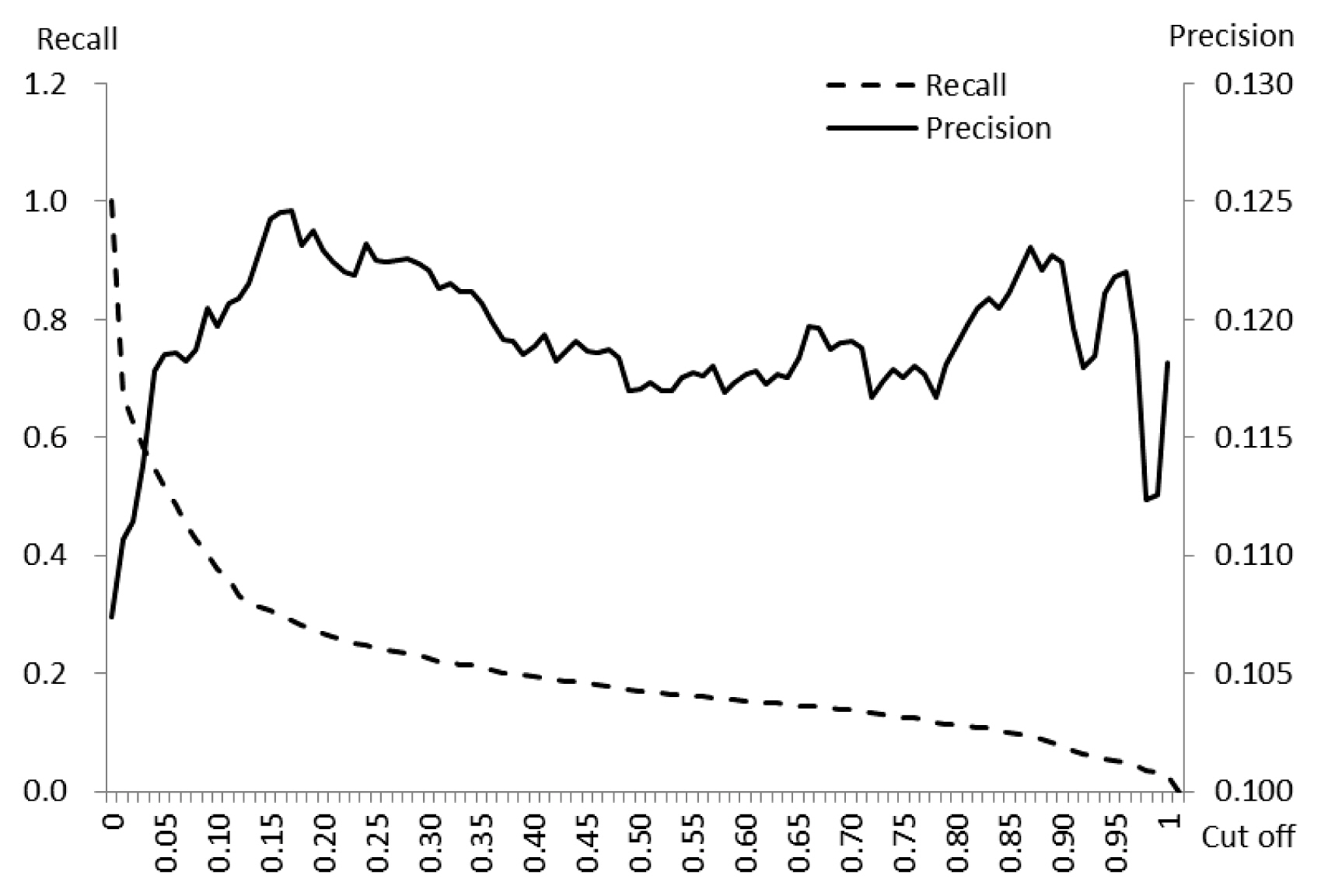
- The precision fluctuated between a minimum 0.107 and a maximum 0.125 (1st quartile 0.118, median 0.119, 3rd quartile 0.122) according to the 101 different cut-offs of predicted risk values. Neither trend of increase nor decrease was detected for precision according to the predicted value of the risk. Meanwhile, as the cut-off increased, the values of recall gradually decreased (Figure 2).
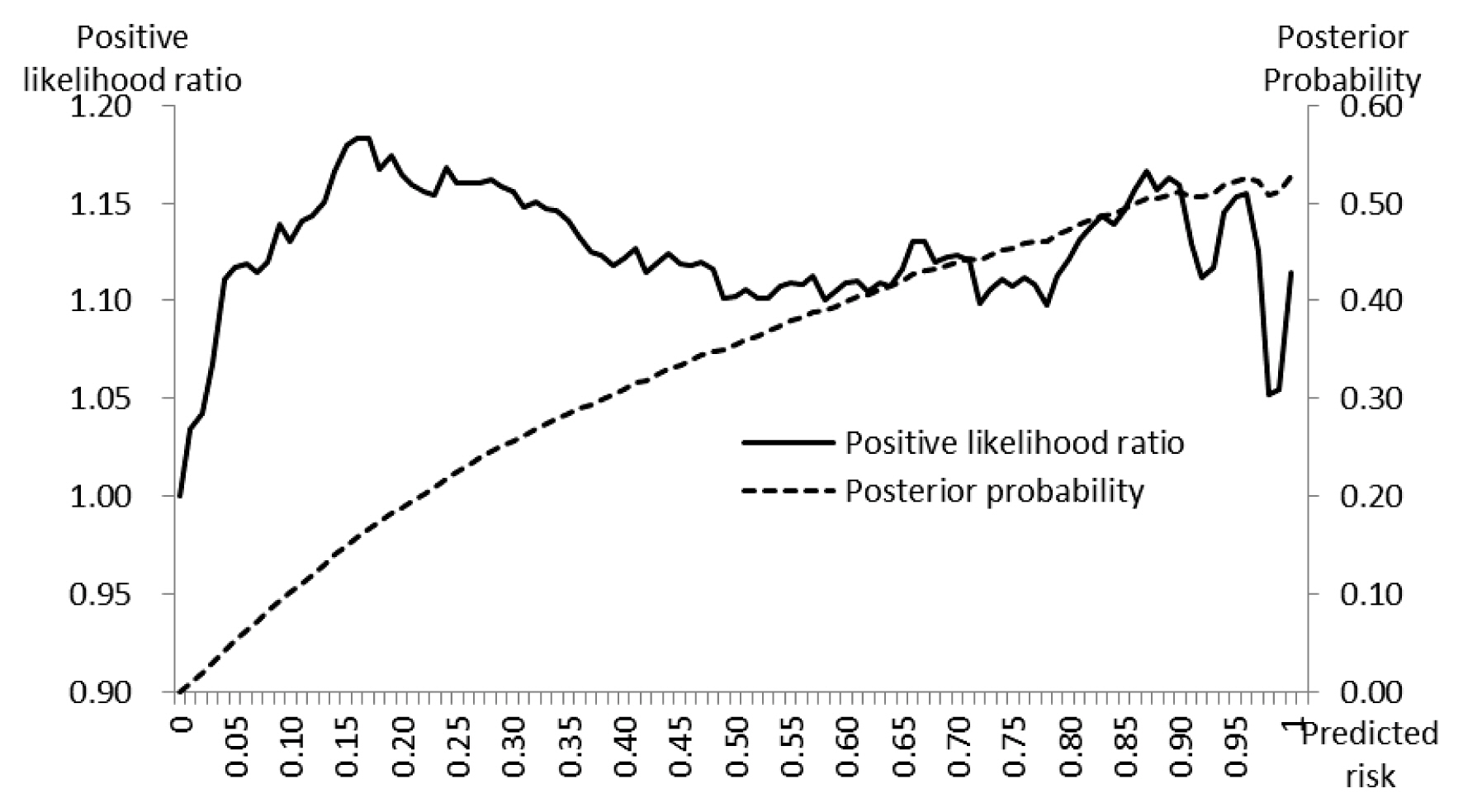
- The range of PLR+ was narrow, it varied from 1.000 (minimum) to 1.183 (maximum), with the first, second (median) and the third quartiles 1.110, 1.123, and 1.150, respectively. The posterior probabilities increased from 0.010 to 0.527 with the increase of cut-off values and it showed a linear pattern with the increase of predicted risk values (Figure 3).
- 3. Application to the 2017/2018 epidemic of HPAI
- The risk assessment program was applied during the HPAI epidemic from November 2017 to March 2018. In association with 22 confirmed outbreaks, a total of 1,217 predictions were generated for 840 poultry farms. Twenty predictions (1.64%) on 11 farms (1.31%: broiler ducks 7, breeder ducks 2, layer hens 2) identified the “outbreak” farms. The predicted mean values of risks were higher for “outbreak” farms (0.25 ± 0.17) than “non-outbreak” farms (0.17 ± 0.29), but the difference was not statistically significant (p = 0.21). However, the predicted values of risk were statistically significantly higher when the related farms were the same farm type as the “outbreak” farms (0.21 ± 0.10, 698 predictions) than a different type of farm (0.12 ± 0.24, 519 predictions).
Results
- This study describes the first risk assessment program in Korea for animal diseases using the deep learning MLP method. This model is a category of DNN, built on the perceptron theory with multiple hidden layers. DNN is the most popular branch of the black-box model, in which parameters are free from the physical boundary of having specific biological meaning [14].
- The core of deep learning is learning from data in order to best predict unobserved data [5,15]. In the MLP model used in this study, the relationship between visit records of vehicles and the status of outbreaks, was developed using a training dataset and was applied to a test dataset (which was not available during the training step), to assign values (probability) of risk to target farms. This process is cross-validation [15]. The validation of the risk model must be performed to assess discrimination (to know whether estimated risks are different for farms with and without outbreaks) and calibration (to measure the agreement between the predicted risks and observed event rates) [16,17]. The model used in this study proved its capacity to discriminate by assigning significantly higher risk values to “outbreak” farms than non-“outbreak” farms. In this study, precision, PLR+, and PPO+ were applied to express model performance. Quartile values of precision between 0.118 (1st) and 0.122 (3rd) signified that amongst predictions having risk values above the cutoff, approximately 12% had outbreaks (Figure 2). In addition, the quartile values PLR+ 1.110 (1st) and 1.150 (3rd) meant that farms which had predicted risk values of higher than the cut-off, had approximately a 1.2 times higher chance of having an outbreak (Figure 3). The PLR+ calibrates how many times more likely the “outbreak” farms are to have the higher predicted risk than “non-outbreak” farms. In conjunction with prior probability and likelihood ratio, the posterior probability was extracted [13]. The highest value of PPO+ 0.5 indicated the probability of outbreak reached 50% after having comprehensively considered the available information (Figure 3).
- Generalization capacity refers to how well the model can work when it is used with data that it has never been exposed to before. In the case of low generalization, over-fitting and subsequently poor predictive performance can be caused by over-training [18]. The generalization capacity of the model was tested by applying it to the epidemic of 2017/2018. Higher risks were predicted for “outbreak” farms than “non-outbreak” farms in the real epidemic. The developed risk assessment model was employed during the epidemics of the 2016/2017 (pilot version) and the 2017/2018 (complementary version). This application induced preventive control measures on the related farms, and consequently a substantial proportion of farms at high risk avoided the outbreak. Therefore, the possibility of a decreasing level of agreement as a result of preemptive control measures being taken in high-risk farms should not be excluded when interpreting calibrations.
Discussion
- This study presented a risk assessment model for HPAI at farm level, developed with a deep learning MLP method. In order to reduce the time required for data processing and improve the quality of results, our model is linked to the KAHIS. The summary of predicted risk was displayed at the websites of the Ministry of Agriculture, Forestry, and Rural Affairs [19] and the APQA [20]. Detailed information was communicated to the national and regional animal health authorities through an official document release system. Our risk assessment model enhanced risk management activities by enabling preemptive control measures to prevent the spread of diseases.
Conclusion
- Details of the explanatory variables included in the risk assessment model.
Supplementary Information
Supplementary Table 1
-
Acknowledgements
- This study was funded by the Animal and Plant Quarantine Agency (R&D Project no.: I-1543068-2019-20-01).
Acknowledgments
- 1. Germeraad EA, Sanders P, Hagenaars TJ, et al. Virus shedding of avian influenza in poultry: A systematic review and meta-analysis. Viruses 2019;11(9). 812PMID: 10.3390/v11090812. PMID: 6784017.ArticlePubMedPMC
- 2. Alexander DJ. A review of avian influenza in different bird species. Vet Microbiol 2000;74(1–2). 3−13. PMID: 10.1016/S0378-1135(00)00160-7. PMID: 10799774.ArticlePubMed
- 3. Rossi G, Smith RL, Pongolini S, et al. Modelling farm-to-farm disease transmission through personnel movements: from visits to contacts, and back. Sci Rep 2017;7:2375PMID: 10.1038/s41598-017-02567-6. PMID: 28539663. PMID: 5443770.ArticlePubMedPMCPDF
- 4. Seo J, Park H, Han KH, et al. Development of mathematical model on regionalization using records of livestock related vehicles for control strategy of highly pathogenic avian influenza. J Prev Vet Med 2017;41:180−5. [in Korean]. PMID: 10.13041/jpvm.2017.41.4.180.Article
- 5. Kashyap H, Afzal Ahmed H, Hoque N, et al. [Preprint]. Big data analytics in bioinformatics: A machine learning perspective 2015 Available from: https://arxiv.org/abs/1506.05101.
- 6. Animal and Plant Quarantine Agency [Internet]. Korea Animal Health Integrated System 2019 [cited 2019 Dec 1]. Available from: http://kahis.go.kr.
- 7. Alexander DJ, Manvell RJ, Irvine R, et al. Overview of incursions of Asian H5N1 subtype highly pathogenic avian influenza virus into Great Britain, 2005–2008. Avian Dis 2010;54(1 Suppl). 194−200. PMID: 10.1637/8833-040209-Reg.1. PMID: 20521632.ArticlePubMed
- 8. Ministry of Agriculture, Forestry and Rural Affairs [Internet]. Standard operation procedure of Highly Pathogenic Avian Influenza in Korea 2018 [cited 2019 Oct 10]. Available from http://www.law.go.kr/admRulLsInfoP.do?ADMRULSEQ=2100000182856.
- 9. Samolov A, Dragovic S, Dakovic M, et al. Analysis of (7) Be behaviour in the air by using a multilayer perceptron neural network. J Environ Radioact 2014;137:198−203. PMID: 10.1016/j.jenvrad.2014.07.016. PMID: 25106024.ArticlePubMed
- 10. Kim DH. Forecasting of social-economic parameters using neural networks. Bull Yosu Nat Univ 2001;16:171−6. [in Korean].
- 11. Lipton ZC, Elkan C, Naryanaswamy B. Optimal thresholding of classifiers to maximize F measure. Mach Learn Knowl Discov Databases 2014;8725:225−39. PMID: 10.1007/978-3-662-44851-9_15.ArticlePubMedPMC
- 12. Akobeng AK. Understanding diagnostic tests 1: sensitivity, specificity, and predictive values. Acta Paediatr 2007;96(3). 338−41. PMID: 10.1111/j.1651-2227.2006.00180.x. PMID: 17407452.ArticlePubMed
- 13. Akobeng AK. Understanding diagnostic tests 2: Likelihood ratios, pre- and post-test probabilities, and their use in clinical practice. Acta Paediatr 2007;96(4). 487−91. PMID: 10.1111/j.1651-2227.2006.00179.x. PMID: 17306009.ArticlePubMed
- 14. Schmidhuber J. Deep learning in neural networks: An overview. Neural Netw 2015;61:85−117. PMID: 10.1016/j.neunet.2014.09.003.ArticlePubMed
- 15. Morota G, Ventura RV, Silva FF, et al. Big data analytics and precision animal agriculture symposium: Machine learning and data mining advance predictive big data analysis in precision animal agriculture. J Anim Sci 2018;96(4). 1540−50. PMID: 10.1093/jas/sky014. PMID: 29385611. PMID: 6140937.ArticlePubMedPMCPDF
- 16. Van Hoorde K, Van Huffel S, Timmerman D, et al. A spline-based tool to assess and visualize the calibration of multiclass risk predictions. J Biomed Inform 2015;54:283−93. PMID: 10.1016/j.jbi.2014.12.016. PMID: 25579635.ArticlePubMed
- 17. Wynants L, Collins GS, Van Calster B. Key steps and common pitfalls in developing and validating risk models. BJOG 2017;124(3). 423−32. PMID: 10.1111/1471-0528.14170.ArticlePubMed
- 18. Lancashire LJ, Lemetre C, Ball GR. An introduction to artificial neural networks in bioinformatics application to complex microarray and mass spectrometry datasets in cancer studies. Brief Bioinform 2009;10(3). 315−29. PMID: 10.1093/bib/bbp012. PMID: 19307287.ArticlePubMedPDF
- 19. Ministry of Agriculture, Forestry and Rural Affairs [Internet]. Information sheet on risk of highly pathogenic avian influenza estimated with big data model 2019 [cited 2019 Dec 1]. Available from: http://www.mafra.go.kr/FMD-AI/641/subview.do.
- 20. Animal and Plant Quarantine Agency [Internet]. Information sheet on risk of highly pathogenic avian influenza estimated with big data model 2019 [cited 2019 Dec 1]. Available from: http://www.qia.go.kr/animal/prevent/listwebQiaCom.do?type=2_33&clear=1.
References
Figure 1
Distribution of risks of HPAI for 23,174 evaluations at the farm level in association with 544 “outbreak” farms from January 2014 to April 2017, predicted by the selected multilayer perceptron model: Comparison by outbreak status of receiver farms.
HPAI = highly pathogenic avian influenza.


Figure 2Recall and precision of the risk predicted by the assessment model, according to a series of cut-off values.
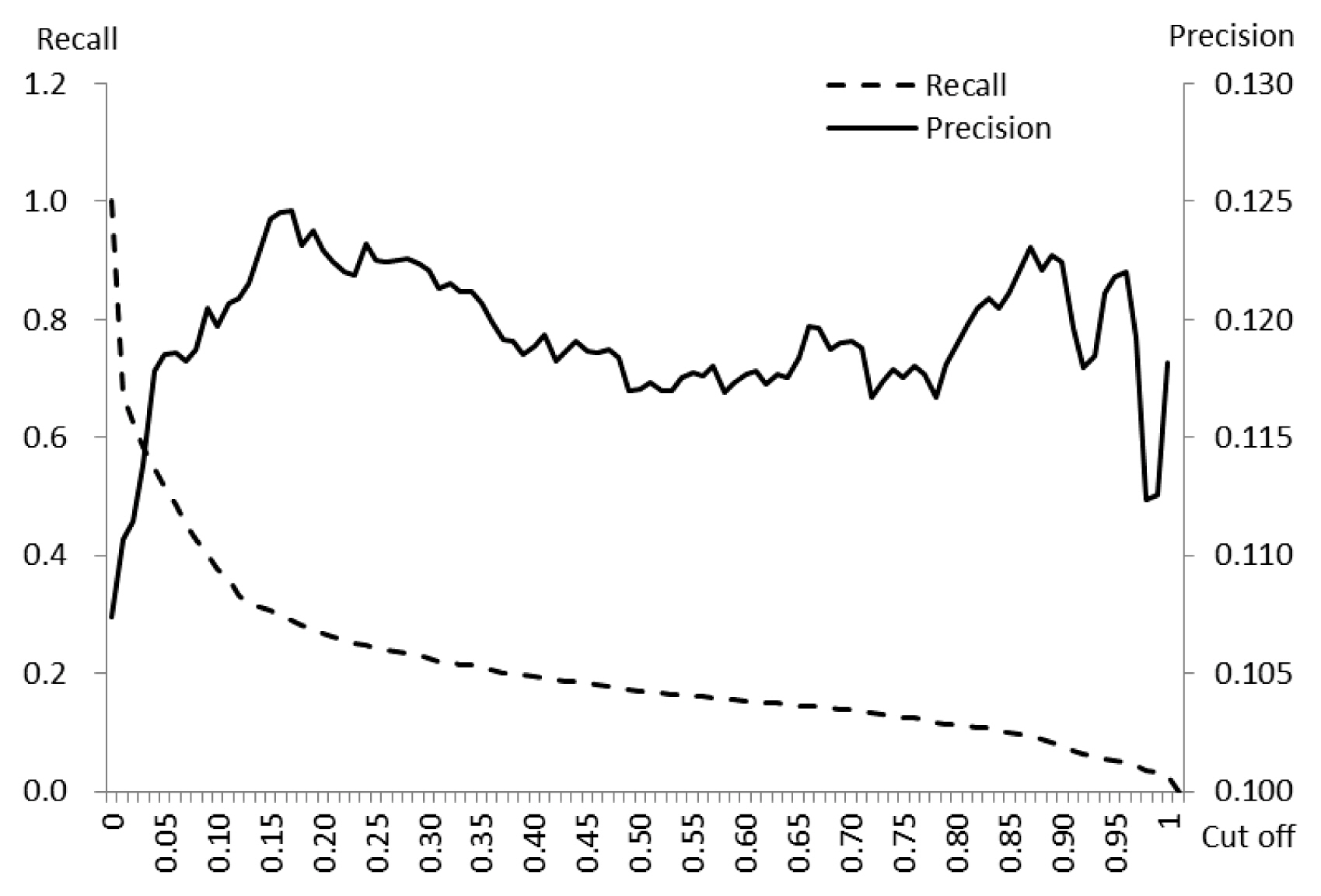

Figure 3Positive likelihood ratio and posterior probability estimated using risk predicted by the risk assessment model.


Table 1Visit records and dangerous contacts generated by livestock-related vehicles during the epidemics of HPAI in Korea from 2014 to 2017.
| Category of use for the vehicle | Total visit records | Dangerous contacts | |||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| No. of vehicles | No. of visits | No. of vehicles | No. of visits | Attack rate (%) by vehicles | |||
|
|
|||||||
| 25th | 50th | 75th | |||||
| Feed | 11,096 | 22,230 | 173 | 1,571 | 8.7 | 14.3 | 27.3 |
|
|
|||||||
| Live animal | 9,110 | 12,464 | 140 | 590 | 6.7 | 10.5 | 22.4 |
|
|
|||||||
| Egg | 916 | 2,549 | 61 | 446 | 33.3 | 50.0 | 66.7 |
|
|
|||||||
| Husk | 976 | 1,727 | 29 | 209 | 16.7 | 25.0 | 40.0 |
|
|
|||||||
| Consultant | 914 | 1,382 | 14 | 79 | 16.9 | 28.9 | 38.3 |
|
|
|||||||
| Veterinary pharmaceuticals | 740 | 1,186 | 6 | 35 | 6.6 | 10.8 | 12.3 |
|
|
|||||||
| Livestock compost | 343 | 984 | 10 | 159 | 18.6 | 50.0 | 72.9 |
|
|
|||||||
| Manure | 280 | 731 | 4 | 105 | 21.3 | 29.2 | 47.9 |
|
|
|||||||
| Veterinarian | 434 | 636 | 4 | 13 | 12.7 | 17.5 | 20.6 |
|
|
|||||||
| Others | 9,534 | 14,137 | 1* | 1 | - | 9.1 | - |
|
|
|||||||
| Total | 34,343 | 58,026 | 442 | 3,208 | 9.1 | 17.2 | 36.4 |
Figure & Data
References
Citations
Citations to this article as recorded by 

- Big data-based risk assessment of poultry farms during the 2020/2021 highly pathogenic avian influenza epidemic in Korea
Hachung Yoon, Ilseob Lee, Hyeonjeong Kang, Kyung-Sook Kim, Eunesub Lee, Mathilde Richard
PLOS ONE.2022; 17(6): e0269311. CrossRef - Artificial Intelligence Models for Zoonotic Pathogens: A Survey
Nisha Pillai, Mahalingam Ramkumar, Bindu Nanduri
Microorganisms.2022; 10(10): 1911. CrossRef


 PubReader
PubReader ePub Link
ePub Link Cite
Cite