Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 9(6); 2018 > Article
-
Original Article
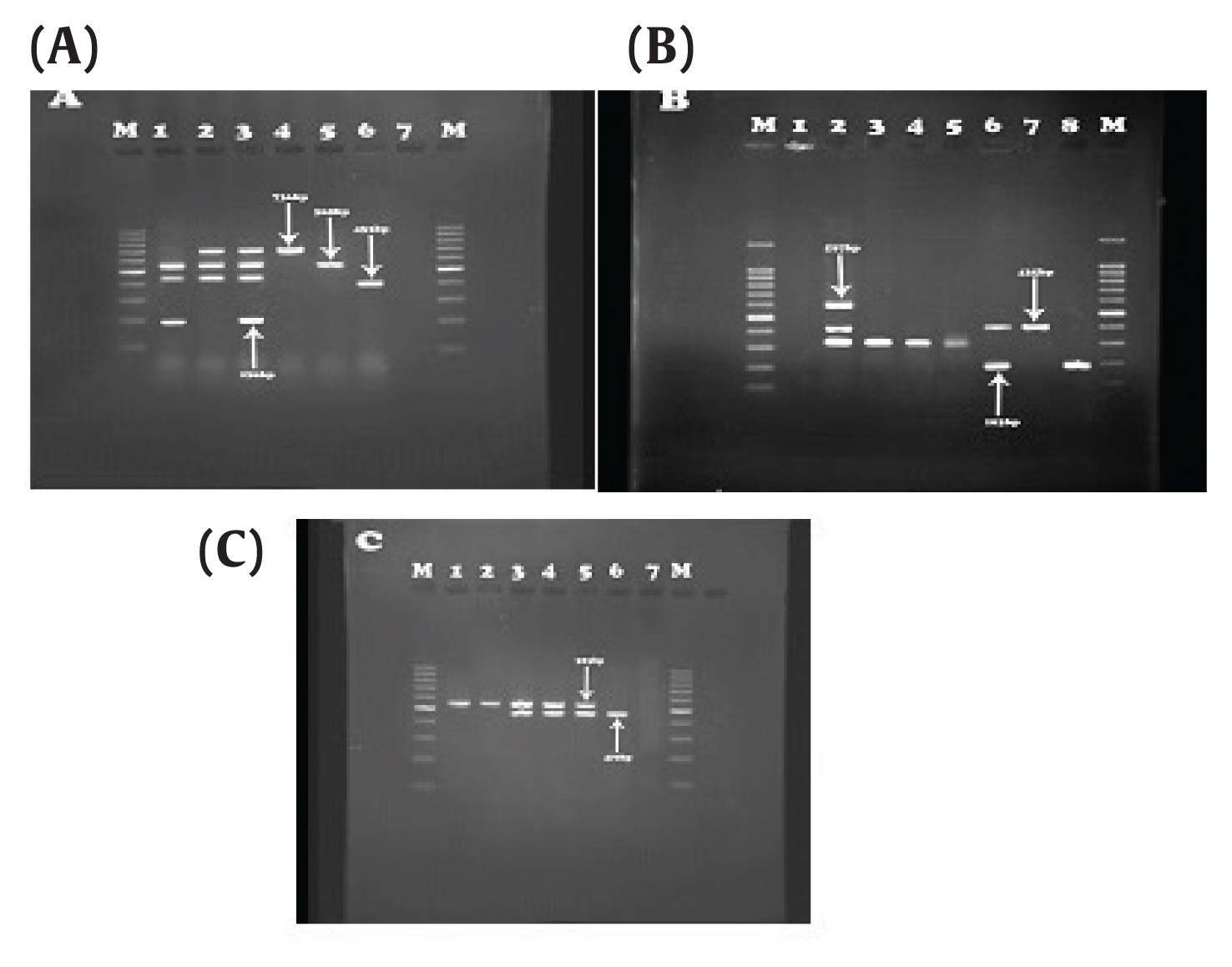
Association between Beta-lactam Antibiotic Resistance and Virulence Factors in AmpC Producing Clinical Strains ofP. aeruginosa - Sanaz Dehbashia, Hamed Tahmasebib, Mohammad Reza Arabestania
-
Osong Public Health and Research Perspectives 2018;9(6):325-333.
DOI: https://doi.org/10.24171/j.phrp.2018.9.6.06
Published online: December 31, 2018
aDepartment of Microbiology, Faculty of Medicine, Hamadan University of Medical Sciences, Hamadan, Iran
bDepartment of Microbiology, Faculty of Medicine, Zahedan University of Medical Sciences, Zahedan, Iran
- *Corresponding author: Mohammad Reza Arabestani, Department of Microbiology, Faculty of Medicine, Brucellosis research center, Hamadan University of Medical Sciences, Hamadan, Iran, E-mail: mohammad.arabestani@gmail.com
• Received: July 10, 2018 • Revised: October 16, 2018 • Accepted: October 22, 2018
Copyright ©2018, Korea Centers for Disease Control and Prevention
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Figure & Data
References
Citations
Citations to this article as recorded by 

- Comparative bacteriome and antibiotic resistome analysis of water and sediment of the Ganga River of India
Ankita Srivastava, Digvijay Verma
World Journal of Microbiology and Biotechnology.2023;[Epub] CrossRef - Prevalence of different virulence factors and their association with antimicrobial resistance among Pseudomonas aeruginosa clinical isolates from Egypt
Eva A. Edward, Marwa R. El Shehawy, Alaa Abouelfetouh, Elsayed Aboulmagd
BMC Microbiology.2023;[Epub] CrossRef - Molecular epidemiology and collaboration of siderophore-based iron acquisition with surface adhesion in hypervirulent Pseudomonas aeruginosa isolates from wound infections
Hamed Tahmasebi, Sanaz Dehbashi, Mona Nasaj, Mohammad Reza Arabestani
Scientific Reports.2022;[Epub] CrossRef - Decoding Genetic Features and Antimicrobial Susceptibility of Pseudomonas aeruginosa Strains Isolated from Bloodstream Infections
Tomasz Bogiel, Dagmara Depka, Mateusz Rzepka, Agnieszka Mikucka
International Journal of Molecular Sciences.2022; 23(16): 9208. CrossRef - Prevalence of the Genes Associated with Biofilm and Toxins Synthesis amongst the Pseudomonas aeruginosa Clinical Strains
Tomasz Bogiel, Dagmara Depka, Mateusz Rzepka, Joanna Kwiecińska-Piróg, Eugenia Gospodarek-Komkowska
Antibiotics.2021; 10(3): 241. CrossRef - A Comprehensive Study of the Relationship between the Production of β-Lactamase Enzymes and Iron/Siderophore Uptake Regulatory Genes in Clinical Isolates of Acinetobacter baumannii
Mahyar Porbaran, Hamed Tahmasebi, MohammadReza Arabestani, Joseph Falkinham
International Journal of Microbiology.2021; 2021: 1. CrossRef - Regulation of virulence and β-lactamase gene expression in Staphylococcus aureus isolates: cooperation of two-component systems in bloodstream superbugs
Sanaz Dehbashi, Hamed Tahmasebi, Behrouz Zeyni, Mohammad Reza Arabestani
BMC Microbiology.2021;[Epub] CrossRef -
New approach to identify colistin‐resistant
Pseudomonas aeruginosa
by high‐resolution melting curve analysis assay
H. Tahmasebi, S. Dehbashi, M.R. Arabestani
Letters in Applied Microbiology.2020; 70(4): 290. CrossRef - Resistant Pseudomonas aeruginosa carrying virulence genes in hospitalized patients with urinary tract infection from Sanandaj, west of Iran
Safoura Derakhshan, Aslan Hosseinzadeh
Gene Reports.2020; 20: 100675. CrossRef - Prevalence and molecular typing of Metallo-β-lactamase-producing Pseudomonas aeruginosa with adhesion factors: A descriptive analysis of burn wounds isolates from Iran
Hamed Tahmasebi, Sanaz Dehbashi, Mohammad Yousef Alikhani, Mahyar Porbaran, Mohammad Reza Arabestani
Gene Reports.2020; 21: 100853. CrossRef - Co-harboring of mcr-1 and β-lactamase genes in Pseudomonas aeruginosa by high-resolution melting curve analysis (HRMA): Molecular typing of superbug strains in bloodstream infections (BSI)
Hamed Tahmasebi, Sanaz Dehbashi, Mohammad Reza Arabestani
Infection, Genetics and Evolution.2020; 85: 104518. CrossRef - Relationship between Biofilm Regulating Operons and Various Β-Lactamase Enzymes: Analysis of the Clinical Features of Infections caused by Non-Fermentative Gram-Negative Bacilli (Nfgnb) from Iran
Mahyar Porbaran, Reza Habibipour
Journal of Pure and Applied Microbiology.2020; 14(3): 1723. CrossRef - Carbapenem-Resistant Pseudomonas aeruginosa Strains-Distribution of the Essential Enzymatic Virulence Factors Genes
Tomasz Bogiel, Małgorzata Prażyńska, Joanna Kwiecińska-Piróg, Agnieszka Mikucka, Eugenia Gospodarek-Komkowska
Antibiotics.2020; 10(1): 8. CrossRef - Biofilm Formation and β-lactamase Enzymes: A Synergism Activity in Acinetobacter baumannii Isolated from Wound Infection
Mahyar Porbaran, Reza Habibipour
Journal of Advances in Medical and Biomedical Rese.2019; 27(125): 34. CrossRef


 PubReader
PubReader Cite
Cite