Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 4(6); 2013 > Article
-
Original Article
Proteomic Analysis of Intracellular and Membrane Proteins From Voriconazole-ResistantCandida glabrata - Jae Il Yooa, Hwa Su Kima, Chi Won Choib, Jung Sik Yooa, Jae Yon Yua, Yeong Seon Leea
-
Osong Public Health and Research Perspectives 2013;4(6):293-300.
DOI: https://doi.org/10.1016/j.phrp.2013.10.001
Published online: October 12, 2013
aDivision of Antimicrobial Resistance, Korea National Institute of Health, Osong, Korea
bProteome Research Team, Korea Basic Science Institute, Daejeon, Korea
- ∗Corresponding author. yslee07@nih.go.kr
© 2013 Published by Elsevier B.V. on behalf of Korea Centers for Disease Control and Prevention.
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
Objectives
- The proteomic analysis of voriconazole resistant Candida glabrata strain has not yet been investigated. In this study, differentially expressed proteins of intracellular and membrane fraction from voriconazole-susceptible, susceptible dose-dependent (S-DD), resistant C. glabrata strains were compared with each other and several proteins were identified.
-
Methods
- The proteins of intracellular and membrane were isolated by disrupting cells with glass bead and centrifugation from voriconazole susceptible, S-DD, and resistant C. glabrata strains. The abundance of expressed proteins was compared using two-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis and proteins showing continuous twofold or more increase or reduction of expression in resistant strains compared to susceptible and S-DD strain were analyzed by liquid chromatography/mass spectrometry-mass spectrometry method.
-
Results
- Of 34 intracellular proteins, 15 proteins showed expression increase or reduction (twofold or more). The identified proteins included regulation, energy production, carbohydrate transport, amino acid transport, and various metabolism related proteins. The increase of expression of heat shock protein 70 was found. Among membrane proteins, 12, 31 proteins showed expression increase or decrease in the order of susceptible, S-DD, and resistant strains. This expression included carbohydrate metabolism, amino acid synthesis, and response to stress-related proteins. In membrane fractions, the change of expression of 10 heat shock proteins was observed, and 9 heat shock protein 70 (Hsp70) showed the reduction of expression.
-
Conclusion
- The expression of Hsp70 protein in membrane fraction is related to voriconazole resistant C. glabrata strains.
- Fungal infection in humans is increasing; Candida species are the most frequently reported organisms. Approximately 95% of all invasive Candida infections are caused by five species: Candida albicans, Candida glabrata, Candida parapsilosis, Candida tropicalis, and Candida krusei [1]. Among the Candida species, C. albicans is the most prevalent in both healthy patients and those with infection [2,3]. Recently, the four non-C. albicans species were found to be more frequently isolated in humans than C. albicans [4]. C. glabrata was the second most common non-C. albicans species in fungemia in the United States and also most commonly recovered from the oral cavities of patients with human immunodeficiency virus [5]. The increase in the number of C. glabrata systemic infections is cause for concern because the high mortality rate associated with C. glabrata fungemia [6]. Because fungal infections are increasing, the use of antifungal agents has correspondingly increased. In particular, fluconazole is a highly effective antifungal agent used for the treatment of candidiasis. Voriconazole is a triazole derivative of fluconazole, and the activity for Candida may be better than that of fluconazole. However, the widespread and prolonged use of fluconazole in recent years has led to the development of drug resistance in Candida species [7,8]. In addition, the resistance of Candida to fluconazole is highly predictive of resistance to voriconazole agent. The observation of cross-resistance in C. glabrata strains receiving fluconazole and voriconazole therapy of C. glabrata in patients with candidemia was reported [9]. The resistant mechanisms to azole antifungal agents have been studied in C. albicans [10–12]. However, C. glabrata has an intrinsic resistant tendency to fluconazole, and the molecular basis for the intrinsically low susceptibility of C. glabrata remains unclear. Several mechanisms of acquired resistance to the azole antifungal agents have been described in C. glabrata. These include upregulation of genes encoding adenosine triphosphate (ATP) binding cassette (ABC) transporters encoded by CDR1 and CDR2 [13]. Overexpression of ERG11, the gene encoding the target of the azole antifungal agents, has also been associated with acquired azole resistance [14]. Recently, proteomic analysis of azole-susceptible and -resistant Candida isolates was accomplished to understand the mechanisms underlying azole antifungal resistance [12,15]. Proteomic analysis has also been used to study the adaptive response of C. albicans to fluconazole and itraconazole [16]. Currently, no proteomic analysis exists for voriconazole resistant C. glabrata strain. So, we analyzed the expression of proteins of voriconazole-susceptible, susceptible dose-dependent (S-DD), and resistant strains to investigate proteins associated with voriconazole resistance.
Introduction
- 2.1 C. glabrata strains and growth conditions
- A total of 56 C. glabrata strains collected from tertiary and nontertiary hospitals were used in this study. We previously reported the results of an antifungal susceptibility test [17]. We selected three C. glabrata strains according to voriconazole susceptibility for a comparative proteomic study. All strains were stored at –80 °C, and prior to the experiment each strain was subcultured twice on sabouraud dextrose agar to ensure viability and purity. For the proteomic experiment, an aliquot of glycerol stock from each strain was diluted in yeast peptone dextrose (YPD; 1% yeast extract, 2% peptone, 1% dextrose) and grown overnight at 30 °C in a shaking incubator. The cultures were diluted to an optical density 0.2 at OD600 in 0.5 L of YPD and grown to the exponential phase of growth.
- 2.2 Cellular protein extraction
- To isolate the cellular proteins, C. glabrata cells were cultured in YPD broth at 30 °C to the exponential phase of growth. Cells were harvested in centrifugation 4000 rpm for 15 minutes. The pellet cells were pooled and washed twice using 50 mM Tris-HCl pH 7.6 buffer solution. The cells were disrupted using 0.45-μm glass beads (Sigma, St. Louis, MO, USA) on ice. After homogenization, the solution was centrifuged twice at 14,000 rpm for 20 minutes. The supernatant was harvested carefully without contaminant similar to a lipid component, and it was freeze dried for further experiment.
- 2.3 Membrane protein extraction
- After an exponential phase of growth, cells were harvested, washed with distilled water, and resuspended in homogenizing buffer (50 mM Tris-HCl, pH 7.5, 2 mM EDTA, 1 mM phenylmethylsulfonylfluoride). After disruption of the cell using the glass bead, cell debris and unbroken cells were removed by centrifugation at 5000 g for 10 minutes. A crude membrane fraction was isolated from the cell-free supernatant by second centrifugation at 30,000 g for 30 minutes. The pellet was washed in GTE buffer (10 mM Tris-HCl, pH 7.0, 0.5 mM EDTA, 20% glucose), resuspended in GTE buffer, and stored at –80 °C. The protein concentration was determined by a micro-Bradford assay using a protein assay kit II (Bio-Rad, Hercules, CA, USA).
- 2.4 Sample preparation and 2-Dimentional Gel Electrophoresis
- The harvested samples were suspended in 0.5 mL of 50 mM Tris buffer containing 7 M urea, 2 M thiourea, 4% [weight/volume (w/v)] CHAPS, and 16 μL protease inhibitor cocktail (Roche Molecular Biochemicals, Indianapolis, IN, USA). The lysates were homogenized and centrifuged at 12,000 × g for 15 minutes. Fifty units of Benzonase (250 units/μL; Sigma) was added to the mixture and suitably stored at −80 °C until use after quantitation by the Bradford method. For 2-DE analysis, pH 3–10 immobilized pH gradient (IPG) strips (Amersham Biosciences, UK, Ltd) were rehydrated in swelling buffer containing 7 M urea, 2 M thiourea, 0.4% (w/v) Dithiothreitol, and 4% (w/v) CHAPS. The protein lysates (500 μg) were cup-loaded into the rehydrated IPG strips using a Multiphor II apparatus (Amersham Biosciences, UK, Ltd) for a total of 57 kVh. The two-dimensional separation was performed on 8–16% (v/v) linear gradient sodium dodecyl sulfate (SDS)-polyacrylamide gels. Following fixation of the gels for 1 hour in a solution of 40% (v/v) methanol containing 5% (v/v) phosphoric acid, the gels were stained with Colloidal Coomassie Blue G-250 solution for 5 hours. The gels were destained in 1% (v/v) acetic acid for 4 hours and then imaged using a GS-710 imaging calibrated densitometer (Bio-Rad).
- Protein spot detection and two-dimensional pattern matching were carried out using ImageMasterTM 2D Platinum software (Amersham Biosciences, UK, Ltd). For comparison of protein spot densities between control and treated samples, more than 20 spots throughout all gels were correspondingly landmarked and normalized. The quantified spots of candidate proteins were compared with the aid of histograms. For ensuring the reproducibility of 2DE experiments, each sample was analyzed in duplicate.
- 2.5 In-gel protein digestion
- Protein bands of interest were excised and digested in-gel with sequencing grade, modified trypsin (Promega, Madison, WI, USA). In brief, each protein spot was excised from the gel, placed in a polypropylene tube, and washed four to five times with 150 μL of 1:1 acetonitrile/25 mM ammonium bicarbonate, pH 7.8. The gel was dried in a Speedvac concentrator, and then rehydrated in 30 μL of 25 mM ammonium bicarbonate, pH 7.8, containing 20 ng of trypsin. After incubation at 37 °C for 20 hours, the liquid was transferred to a new tube. Tryptic peptides remaining in the gel matrix were extracted for 40 min at 30 °C with 20 μL of 50% (v/v) aqueous acetonitrile containing 0.1% (v/v) formic acid. The combined supernatants were evaporated in a Speedvac concentrator and dissolved in 8 μL of 5% (v/v) aqueous acetonitrile solution containing 0.1% (v/v) formic acid for mass spectrometric analysis.
- 2.6 Identification of proteins by liquid chromatograph/tandem mass spectrometry
- The resulting tryptic peptides were separated and analyzed using reversed phase capillary high-performance liquid chromatography (HPLC) directly coupled to a Finnigan LCQ ion trap mass spectrometer [liquid chromatography-tandem mass spectrometry (LC-MS/MS)]. A 0.1 × 20 mm trapping and a 0.075 × 130 mm resolving column were packed with Vydac 218 MS low trifluoroacetic acid C18 beads (5 μm in size, 300 Å in pore size; Vydac, Hesperia, CA, USA) and placed in-line. Next, the peptides were bound to the trapping column for 10 minutes with 5% (v/v) aqueous acetonitrile containing 0.1% (v/v) formic acid, then the bound peptides were eluted with a 50-minute gradient of 5-80% (v/v) acetonitrile containing 0.1% (v/v) formic acid at a flow rate of 0.2 μL/min. For tandem mass spectrometry, a full mass scan range mode was m/z = 450–2000 Da. After determination of the charge states of an ion on zoom scans, product ion spectra were acquired in MS/MS mode with relative collision energy of 55%. The individual spectra from MS/MS were processed using the TurboSEQUEST software (Thermo Quest, San Jose, CA). The generated peak list files were used to query either the MSDB database or National Center for Biotechnology Information (NCBI) using the MASCOT program (http://www.matrixscience.com). Modifications of methionine and cysteine, peptide mass tolerance at 2 Da, MS/MS ion mass tolerance at 0.8 Da, allowance of missed cleavage at 2, and charge states (+1, +2, and +3) were taken into account. Only significant hits as defined by MASCOT probability analysis were initially considered.
Materials and methods
- 3.1 Strains
- Among the C. glabrata strains, voriconazole susceptible strain [C. glabrata I-49, minimum inhibitory concentration (MIC) 0.5 μg/mL], S-DD strain (C. glabrata D-54, MIC 2 μg/mL) and resistant strain (C. glabrata D-91, MIC 4 μg/mL) were selected. All strains were isolated from blood specimen of patients.
- 3.2 Expression of intracellular proteins and identification
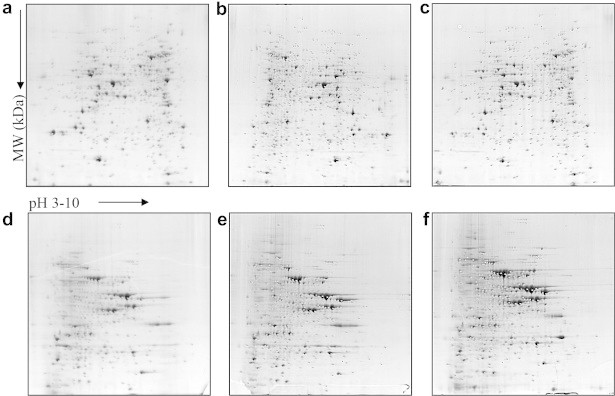
- The two-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis (2D-SDS PAGE) gels are shown in Figure 1. The profiling of 459 intracellular proteins was detected in three strains. Of the total proteins, 38 proteins having abundance ratios of twofold or more showed continuous increase of expression from susceptible and S-DD to resistant strain. In addition, 34 proteins were identified by LC-MS/MS (Table 1). The 15 proteins showing decrease of expression from susceptible and S-DD to resistant strain were also identified. Among the identified proteins, aldehyde dehydrogenase family, serine hydroxymethyltransferase, acetolactate synthase, heat shock protein, pyruvate kinase, potassium efflux protein, isocitrate dehydrogenase, and other proteins showed increased expression. Expression was decreased in proteins such as glycerol-3-phosphate dehydrogenase, ATP synthase, acetyl-coA hydrolase, oxidoreductase, and malate dehydrogenases (Table 1). Among the proteins for which expression was decreased, phosphoglycerate kinase protein showed the largest decreased expression, at 9.09 times reduction of expression. The identified proteins, classified according to their function, are summarized in Table 2. The functional category showed that the identified proteins were cell regulation, energy production, carbohydrate transport, amino acid transport, and various metabolism-related proteins.
- 3.3 Expression of membrane proteins and identification
- A total of 329 membrane proteins were resolved by 2D gel electrophoresis. Of the 17 spots (differential ratio twofold or more) for which expression was increased, 12 proteins were identified. The identified proteins showed enolase, heat shock protein 70, pyruvate kinase, cysteine synthase, pyruvate decarboxylase, pyrophosphate requiring enzyme, regulatory modules in signal transduction, and phosphoglycerate kinase (Table 3). Among the identified proteins, phosphate requiring enzymes showed the most increased expression (3.66 times). Enolase and phosphoglycerate kinase proteins also showed 2.69 and 2.77 times increased expression, respectively. Thirty-seven spots showed decreased expression in the order of susceptible, S-DD, and resistant strains. Among the 37 spots, 31 proteins were identified. The identified membrane proteins included heat shock protein 70, aldehyde dehydrogenase, nicotinamide adenine dinucleotide phosphate-glutamate dehydrogenase, phosphoglycerate mutase I, glutamine aminotransferase, superoxide dismutase, Stm1 protein, phosphoglycerate kinase, and others. A total of 12 heat shock proteins were observed and heat shock protein 70 was 11. In addition, 9 heat shock protein 70 showed the deceased expression in resistant strain compared to susceptible and S-DD strain. The identified membrane proteins were classified into carbohydrate metabolism, amino acid synthesis, and response to stress-related proteins (Table 4).
Results
- C. glabrata is a major opportunistic fungal pathogen of humans and also part of the gastrointestinal microflora in many healthy human beings [1]. The most effective classes of antifungal agents used to treat C. glabrata infections are the azoles agents, specifically fluconazole and voriconazole [9]. However, the occurrence of azole-resistant strains resulted in a difficulty of treatment. Currently, the available information of voriconazole resistance in protein levels is sparse. In this study, we compared the expression changes of proteins using the voriconazole susceptible, S-DD, and resistant strains. The results of proteomic analysis showed the tendency of expression increase (38 proteins) was observed in intracellular fractions of resistant strain compared to membrane fraction of susceptible and S-DD strain (17 proteins). The membrane fraction of resistant strain had the tendency of expression decrease (37 proteins) compared to intracellular fraction of susceptible and S-DD strains (18 proteins). The results indicated that the metabolism process is continuously increased from voriconazole susceptible to S-DD, resistant strain but the biochemical reaction may be decreased in membrane fraction to endure the antifungal stress environment. Among the identified proteins, heat shock protein was observed in various spots of intracellular and membrane fractions. Usually, heat shock protein is known as a stress and response related protein. In this study, the expression increase of heat shock protein in intracellular proteins of voriconazole resistant strain was observed in three spots, but 9 heat shock 70 protein showed decreased expression in membrane proteins. This finding indicated that heat shock protein 70 is related to voriconazole resistance. Among the C. albicans triazole resistance mechanisms, the molecular chaperone Hsp90 is known to share a correlation. The Hsp90 protein stabilizes calcineurin, thereby enabling calcineurin-dependent stress responses that are required for triazole tolerance of Candida strains [18]. In this study, the heat shock protein identified most often was Hsp70 protein, and 9 Hsp70 proteins showed a decrease of expression in membrane fraction, but the exact mechanism with voriconazole resistance needs more investigation. Among the identified membrane proteins, expression of DnaK and Stml protein was reduced in voriconazole resistant strains compared with S-DD and susceptible strains. These proteins are related to protein posttranslation modification and apoptosis, respectively. There has been little information of voriconazole resistance in C. glabrata strain, so the proteomic investigation can be useful information for further study.
Discussion
-
Acknowledgements
- This study was supported by an intramural research grant from the Korea Centers for Disease Control and Prevention (grant no: 2006-N44002-00)
Acknowledgment
-
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Article information
- 1. Pfaller M.A., Diekema D.J.. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20(1). 2007 Jan;133−163. PMID: 17223626.ArticlePubMed
- 2. Hooshdaran M.Z., Barker K.S., Hilliard G.M., Kusch H., Morschhauser J., Rogers P.D.. Proteomic analysis of azole resistance in Candida albicans clinical isolates. Antimicrob Agents Chemother 48(7). 2004 Jul;2733−2735. PMID: 15215138.ArticlePubMed
- 3. Samaranayake L.P., Fidel P.L., Naglik J.R.. Fungal infections associated with HIV infection. Oral Dis 8(Suppl. 2). 2002 Jul;151−160. PMID: 12164650.ArticlePubMed
- 4. Krcmery V., Barnes A.J.. Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. J Hosp Infect 50(4). 2002 Apr;243−260. PMID: 12014897.ArticlePubMed
- 5. Melo N.R., Taguchi H., Jorge J.. Oral candida flora from Brazilian human immunodeficiency virus-infected patients in the highly active antiretroviral therapy era. Mem Inst Oswaldo Cruz 99(4). 2004 Jun;425−431. PMID: 15322634.ArticlePubMed
- 6. Tumbarello M., Sanguinetti M., Trecarichi E.M.. Fungaemia caused by Candida glabrata with reduced susceptibility to fluconazole due to altered gene expression: risk factors, antifungal treatment and outcome. J Antimicrob Chemother 62(6). 2008 Dec;1379−1385. PMID: 18782778.ArticlePubMed
- 7. Andes D., Lepak A., Nett J., Lincoln L., Marchillo K.. In vivo fluconazole pharmacodynamics and resistance development in a previously susceptible Candida albicans population examined by microbiologic and transcriptional profiling. Antimicrob Agents Chemother 50(7). 2006 Jul;2384−2394. PMID: 16801416.ArticlePubMed
- 8. Masia C.M., Gutierrez R.F.. Antifungal drug resistance to azoles and polyenes. Lancet Infect Dis 2(9). 2002 Sep;550−563. PMID: 12206971.ArticlePubMed
- 9. Lyon G.M., Karatela S., Sunay S., Adiri Y.. Antifungal susceptibility testing of Candida isolates from the Candida surveillance study. J Clin Microbiol 48(4). 2010 Apr;1270−1275. PMID: 20129963.ArticlePubMed
- 10. Bennett J.E., Izumikawa K., Marr K.A.. Mechanism of increased fluconazole resistance in Candida glabrata during prophylaxis. Antimicrob Agents Chemother 48(5). 2004 May;1773−1777. PMID: 15105134.ArticlePubMed
- 11. Redding S.W., Kirkpatrick W.R., Saville S.. Multiple patterns of resistance to fluconazole in Candida glabrata isolates from a patient with oropharyngeal candidiasis receiving head and neck radiation. J Clin Microbiol 41(2). 2003 Feb;619−622. PMID: 12574256.ArticlePubMed
- 12. Sanguinetti M., Posteraro B., Fiori B., Ranno S., Torelli R., Fadda G.. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob Agents Chemother 49(2). 2005 Feb;668−679. PMID: 15673750.ArticlePubMed
- 13. Vermitsky J.P., Edlind T.D.. Azole resistance in Candida glabrata: coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob Agents Chemother 48(10). 2004 Oct;3773−3781. PMID: 15388433.ArticlePubMed
- 14. Geber A., Hitchcock C.A., Swartz J.E.. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob Agents Chemother 39(12). 1995 Dec;2708−2717. PMID: 8593007.ArticlePubMed
- 15. De Groot P.W., de Boer A.D., Cunningham J.. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot Cell 3(4). 2004 Aug;955−965. PMID: 15302828.ArticlePubMed
- 16. Bruneau J.M., Maillet I., Tagat E.. Drug induced proteome changes in Candida albicans: comparison of the effect of beta(1,3) glucan synthase inhibitors and two triazoles, fluconazole and itraconazole. Proteomics 3(3). 2003 Mar;325−336. PMID: 12627386.ArticlePubMed
- 17. Yoo J.I., Cho C.W., Lee K.M.. National surveillance of antifungal susceptibility of Candida species in South Korean hospital. Med Mycol 47(5). 2009;534−538.
- 18. Cowen L.E., Steinbach W.J.. Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryot Cell 7(5). 2008 May;747−764. PMID: 18375617.ArticlePubMed
References
| Spot | Protein | Molecular mass (Dalton) | pI | Fold change (R/S)a |
|---|---|---|---|---|
| 25 | C1-tetrahydrofolate synthase | 102,203 | 5.98 | 3.09 |
| 27 | Formyltetrahydrofolate synthetase (FTHFS) | 102,203 | 5.98 | 2.06 |
| 79 | ACO1 aconitate hydratase aconitase | 85,429 | 6.78 | 2.53 |
| 90 | Potassium efflux protein KefA | 73,694 | 5.41 | 2.05 |
| 115 | Sphingolipid long-chain base sensory protein | 40,387 | 5.54 | 2.27 |
| 116 | TKL1 transketolase | 73,704 | 6.01 | 2.50 |
| 127 | Heat shock protein 70 | 112,540 | 7.87 | 3.5 |
| 189 | 2.15 | |||
| 540 | 2.13 | |||
| 154 | Acetolactate synthase | 73,300 | 8.55 | 2.41 |
| 202 | LEU4 2-isopropylmalalate synthase | 67,290 | 5.52 | 2.51 |
| 218 | Acetyl-CoA hydrolase/transferase N-terminal domain | 58,541 | 6.16 | 2.38 |
| 228 | Phosphoribosylaminoimidazole carboxylase | 62,672 | 6.95 | 4.42 |
| 238 | Pyruvate kinase | 55,563 | 6.25 | 2.66 |
| 263 | Pyruvate decarboxylase and related thiamine pyrophosphate-requiring enzymes | 61,726 | 5.59 | 3.94 |
| 295 | Aldehyde dehydrogenase family | 55,937 | 5.09 | 2.03 |
| 304 | SES1 seryl-transcription RNA synthetase | 52,775 | 5.8 | 2.19 |
| 319 | Iinosine monophosphate dehydrogenase | 56,969 | 6.69 | 2.86 |
| 396 | Serine hydroxymethyltransferase | 52,271 | 6.74 | 3.1 |
| 397 | 3.6 | |||
| 411 | GDP dissociation inhibitor | 50,582 | 5.66 | 3.02 |
| 476 | Protein with specific affinity for G4 quadruplex nucleic acids | 42,134 | 8.61 | 2.33 |
| 504 | Isocitrate dehydrogenase | 46,728 | 5.23 | 2.52 |
| 505 | spP36046 Saccharomyces cerevisiae YKL195w | 44,592 | 4.45 | 2.78 |
| 507 | Chromosome segregation adenosine triphosphatases (ATPases) | 55,271 | 8.81 | 14.1 |
| 535 | Malate dehydrogenases (MDH) glycosomal and mitochondrial | 39,024 | 6.15 | 2.42 |
| 550 | Cyclophilin_ABH_like | 41,620 | 5.61 | 2.2 |
| 560 | Aspartate/tyrosine/aromatic aminotransferase | 45,608 | 7.2 | 2.73 |
| 576 | Quinone reductase and related Zn-dependent oxidoreductases | 40,823 | 6.01 | 2.24 |
| 603 | Branched-chain aminotransferase | 41,550 | 5.82 | 2.58 |
| 619 | Highly similar to S. cerevisiae YBR249c ARO4 | 38,617 | 6.51 | 2.24 |
| 636 | RPC40 DNA-directed RNA polymerase I | 37,577 | 5.22 | 2.05 |
| 645 | S. cerevisiae YGR080w | 36,175 | 5.02 | 3.49 |
| 777 | Peptidase_S8 (serine proteinase) | 50,008 | 5.75 | 2.25 |
| 107 | Glycerol-3-phosphate dehydrogenase | 43,961 | 5.85 | −2.82 |
| 142 | TKL1 transketolase | 73,704 | 6.01 | −3.57 |
| 321 | F0F1 ATP synthase | 58,485 | 8.99 | −2.46 |
| 454 | Effector domain of the CAP family of transcription factors | 44,936 | 5.92 | −2.14 |
| 570 | Acetyl-CoA hydrolase | −2.53 | ||
| 609 | Oxidoreductases | 46,710 | 5.76 | −2.4 |
| 613 | −3.75 | |||
| 615 | Malate dehydrogenases glycosomal and mitochondrial | 40,487 | 9.18 | −2.14 |
| 625 | Phosphoglycerate kinase | 44,590 | 6.37 | −8.90 |
| 628 | Arginase | 35,061 | 5.27 | −3.13 |
| 633 | Highly similar to spP53252 S. cerevisiae YGR086c | 35,129 | 4.68 | −3.03 |
| 723 | Uncharacterized enzymes related to aldose 1-epimerase | 33,397 | 5.06 | −3.15 |
| 774 | −2.07 | |||
| 909 | Hypothetical protein CAGL0I00616g | 2,183 | 5.37 | −9.09 |
| 912 | ATP synthase D chain, mitochondrial (ATP5H) | 19,918 | 6.64 | −2.73 |
| Spot | Protein | Molecular mass (Dalton) | pI | Fold change (R/S)a |
|---|---|---|---|---|
| 12, 314 | Enolase | 46,710 | 5.76 | 2.69, 2.56 |
| 132, 169 | Hsp70 protein | 6,635 | 5.32 | 2.18, 2.72 |
| 195, 379 | Pyruvate kinase (PK) | 54,572 | 8.26 | 2.13, 2.21 |
| 244 | Cysteine synthase | 55,388 | 5.51 | 2.34 |
| 255, 291 | Pyruvate decarboxylase | 61,726 | 5.59 | 2.12, 2.31 |
| 276 | Pyrophosphate-requiring enzymes | 46,993 | 4.46 | 3.66 |
| 284 | WD40 domain adaptor/regulatory modules in signal transduction | 46,504 | 4.44 | 2.34 |
| 457 | Phosphoglycerate kinase (PGK) | 44,590 | 6.37 | 2.77 |
| 50 | Heat shock protein | 80,983 | 4.82 | −2.43 |
| 119, 149 | Hsp70 protein | 69,469 | 4.96 | −2.43, −4.09 |
| 153, 226 | −2.18, −2.74 | |||
| 357, 552 | −2.54, −3.88 | |||
| 138, 174 | −28.0, −2.37 | |||
| 172 | −3.35 | |||
| 175 | Saccharomyces cerevisiae YLR259c Heat shock protein | 60,351 | 5.14 | −4.77 |
| 229 | Hexokinase | 53,772 | 5.23 | −2.39 |
| 260 | Aldehyde dehydrogenase family | 56,131 | 6.07 | −2.11 |
| 292 | F1 adenosine triphosphate (ATP) synthase beta subunit, nucleotide-binding domain | 54,176 | 5.14 | −2.68 |
| 298 | Nicotinamide adenine dinucleotide phosphate -glutamate dehydrogenase | 49,711 | 5.58 | −3.17 |
| 360 | SAM1 S-adenosylmethionine synthetase | 41,700 | 5.10 | −2.19 |
| 398 | ATPase alpha2,Na/K | 116,305 | 5.41 | −2.56 |
| 462 | N terminal of the Stm1 protein | 29,791 | 9.65 | −6.48 |
| 465 | Adenosine kinase (AK) | 36,250 | 5.23 | −2.05 |
| 548 | Exo-beta-1,3-glucanase | 33,667 | 4.41 | −2.50 |
| 557 | Elongation factor 1 beta (EF1B) guanine nucleotide exchange domain | 22,903 | 4.33 | −3.57 |
| 560 | Predicted epimerase, PhzC/PhzF homolog | 32,286 | 4.98 | −2.35 |
| 578 | Phosphoglycerate mutase 1 | 27,468 | 5.48 | −11.1 |
| 597 | Phosphoglycerate kinase | 18,458 | 7.85 | −2.51 |
| 602 | Mitochondrial ribosomal protein MRP8 | 24,160 | 4.73 | −2.12 |
| 616 | Ribosome antiassociation factor IF6 | 26,367 | 4.52 | −2.62 |
| 629 | TrpR binding protein WrbA | 29,728 | 6.54 | −2.08 |
| 632 | Alcohol dehydrogenase GroES-like domain | 36,721 | 6.21 | −2.25 |
| 645 | Type 1 glutamine amidotransferase (GATase1) | 25,479 | 5.16 | −4.45 |
| 658 | Phosphoglycerate kinase (PGK) | 44,590 | 6.37 | −2,15 |
| 715 | Chain A, yeast Cu, Zn enzyme superoxide dismutase | 15,714 | 5.63 | −7.19 |
Figure & Data
References
Citations

- What ‘Omics can tell us about antifungal adaptation
Gabriela Fior Ribeiro, Eszter Denes, Helen Heaney, Delma S Childers
FEMS Yeast Research.2022;[Epub] CrossRef - Effects of antifungal agents on the fungal proteome: informing on mechanisms of sensitivity and resistance
Rebecca A. Owens, Sean Doyle
Expert Review of Proteomics.2021; 18(3): 185. CrossRef - HPLC-MS identification and expression of Candida drug-resistance proteins from African HIV-infected patients
Pedro M D S Abrantes, Randall Fisher, Patrick J D Bouic, Carole P McArthur, Burtram C Fielding, Charlene W J Africa
AIMS Microbiology.2021; 7(3): 320. CrossRef


 PubReader
PubReader Cite
Cite
