Search
- Page Path
- HOME > Search
- Prevalence, multidrug resistance, and biofilm formation of Vibrio parahaemolyticus isolated from fish mariculture environments in Cat Ba Island, Vietnam
- Kim Cuc Thi Nguyen, Phuc Hung Truong, Hoa Truong Thi, Xuan Tuy Ho, Phu Van Nguyen
- Osong Public Health Res Perspect. 2024;15(1):56-67. Published online February 19, 2024
- DOI: https://doi.org/10.24171/j.phrp.2023.0181
- 752 View
- 45 Download
-
 Graphical Abstract
Graphical Abstract
 Abstract
Abstract
 PDF
PDF 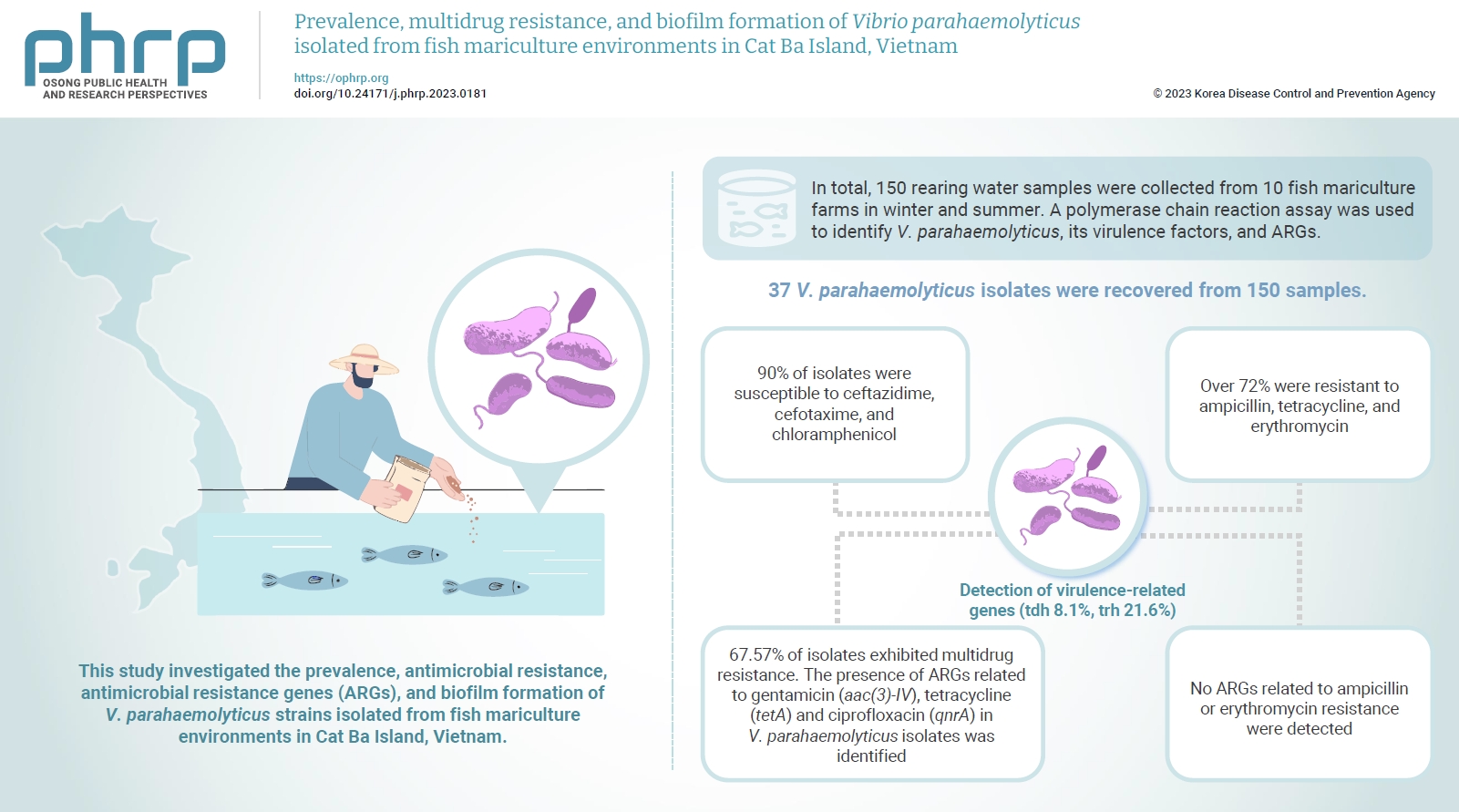
- Objectives
Vibrio parahaemolyticus is a major foodborne pathogen in aquatic animals and a threat to human health worldwide. This study investigated the prevalence, antimicrobial resistance, antimicrobial resistance genes (ARGs), and biofilm formation of V. parahaemolyticus strains isolated from fish mariculture environments in Cat Ba Island, Vietnam. Methods: In total, 150 rearing water samples were collected from 10 fish mariculture farms in winter and summer. A polymerase chain reaction assay was used to identify V. parahaemolyticus, its virulence factors, and ARGs. The antimicrobial resistance patterns and biofilm formation ability of V. parahaemolyticus strains were investigated using the disk diffusion test and a microtiter plate-based crystal violet method, respectively. Results: Thirty-seven V. parahaemolyticus isolates were recovered from 150 samples. The frequencies of the tdh and trh genes among V. parahaemolyticus isolates were 8.1% and 21.6%, respectively. More than 90% of isolates were susceptible to ceftazidime, cefotaxime, and chloramphenicol, but over 72% were resistant to ampicillin, tetracycline, and erythromycin. Furthermore, 67.57% of isolates exhibited multidrug resistance. The presence of ARGs related to gentamicin (aac(3)-IV), tetracycline (tetA) and ciprofloxacin (qnrA) in V. parahaemolyticus isolates was identified. Conversely, no ARGs related to ampicillin or erythromycin resistance were detected. Biofilm formation capacity was detected in significantly more multidrug-resistant isolates (64.9%) than non-multidrug-resistant isolates (18.9%). Conclusion: Mariculture environments are a potential source of antibiotic-resistant V. parahaemolyticus and a hotspot for virulence genes and ARGs diffusing to aquatic environments. Thus, the prevention of antibiotic-resistant foodborne vibriosis in aquatic animals and humans requires continuous monitoring.
- Drug resistance and the genotypic characteristics of rpoB and katG in rifampicin- and/or isoniazid-resistant Mycobacterium tuberculosis isolates in central Vietnam
- Thi Binh Nguyen Nguyen, Thi Kieu Diem Nguyen, Van Hue Trương, Thi Tuyet Ngoc Tran, van Bao Thang Phan, Thi Tuyen Nguyen, Hoang Bach Nguyen, Viet Quynh Tram Ngo, Van Tuan Mai, Paola Molicotti
- Osong Public Health Res Perspect. 2023;14(5):347-355. Published online October 18, 2023
- DOI: https://doi.org/10.24171/j.phrp.2023.0124
- 1,123 View
- 76 Download
-
 Graphical Abstract
Graphical Abstract
 Abstract
Abstract
 PDF
PDF 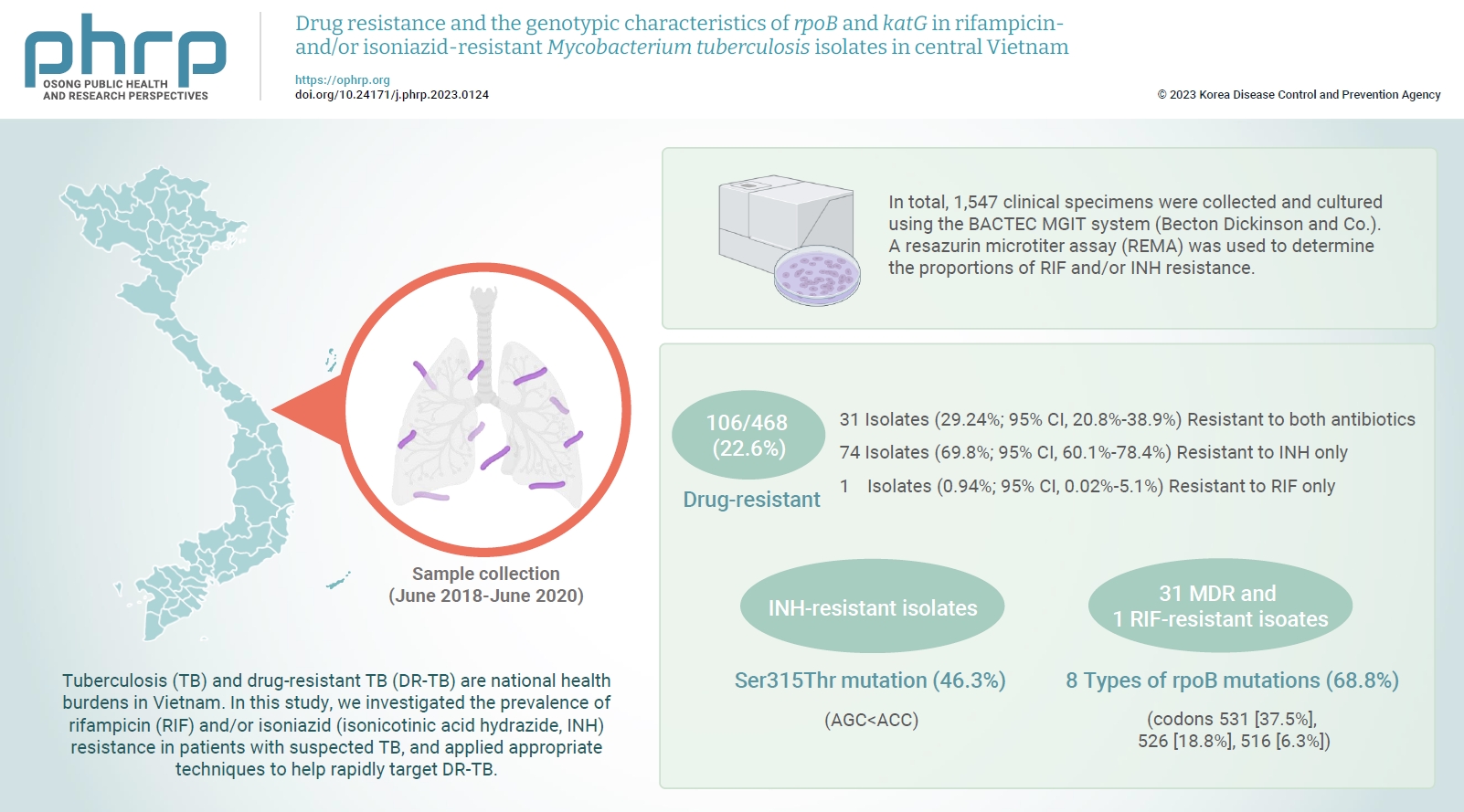
- Objectives
Tuberculosis (TB) and drug-resistant TB (DR-TB) are national health burdens in Vietnam. In this study, we investigated the prevalence of rifampicin (RIF) and/or isoniazid (isonicotinic acid hydrazide, INH) resistance in patients with suspected TB, and applied appropriate techniques to help rapidly target DR-TB. Methods: In total, 1,547 clinical specimens were collected and cultured using the BACTEC MGIT system (Becton Dickinson and Co.). A resazurin microtiter assay (REMA) was used to determine the proportions of RIF and/or INH resistance. A real-time polymerase chain reaction panel with TaqMan probes was employed to identify the mutations of rpoB and katG associated with DR-TB in clinical isolates. Genotyping of the identified mutations was also performed. Results: A total of 468 Mycobacterium tuberculosis isolates were identified using the REMA. Of these isolates, 106 (22.6%) were found to be resistant to 1 or both antibiotics. Of the resistant isolates, 74 isolates (69.8%) were resistant to isoniazid (INH) only, while 1 isolate (0.94%) was resistant to RIF only. Notably, 31 isolates (29.24%) were resistant to both antibiotics. Of the 41 phenotypically INH-resistant isolates, 19 (46.3%) had the Ser315Thr mutation. There were 8 different rpoB mutations in 22 (68.8%) of the RIF-resistant isolates. The most frequently detected mutations were at codons 531 (37.5%), 526 (18.8%), and 516 (6.3%). Conclusion: To help prevent new cases of DR-TB in Vietnam, it is crucial to gain a comprehensive understanding of the genotypic DR-TB isolates.
- Strategies to combat Gram-negative bacterial resistance to conventional antibacterial drugs: a review
- Priyanka Bhowmik, Barkha Modi, Parijat Roy, Antarika Chowdhury
- Osong Public Health Res Perspect. 2023;14(5):333-346. Published online October 18, 2023
- DOI: https://doi.org/10.24171/j.phrp.2022.0323
- 2,122 View
- 169 Download
- 1 Web of Science
- 2 Crossref
-
 Graphical Abstract
Graphical Abstract
 Abstract
Abstract
 PDF
PDF 
- The emergence of antimicrobial resistance raises the fear of untreatable diseases. Antimicrobial resistance is a multifaceted and dynamic phenomenon that is the cumulative result of different factors. While Gram-positive pathogens, such as methicillin-resistant Staphylococcus aureus and Clostridium difficile, were previously the most concerning issues in the field of public health, Gram-negative pathogens are now of prime importance. The World Health Organization’s priority list of pathogens mostly includes multidrug-resistant Gram-negative organisms particularly carbapenem-resistant Enterobacterales, carbapenem-resistant Pseudomonas aeruginosa, and extensively drug-resistant Acinetobacter baumannii. The spread of Gram-negative bacterial resistance is a global issue, involving a variety of mechanisms. Several strategies have been proposed to control resistant Gram-negative bacteria, such as the development of antimicrobial auxiliary agents and research into chemical compounds with new modes of action. Another emerging trend is the development of naturally derived antibacterial compounds that aim for targets novel areas, including engineered bacteriophages, probiotics, metal-based antibacterial agents, odilorhabdins, quorum sensing inhibitors, and microbiome-modifying agents. This review focuses on the current status of alternative treatment regimens against multidrug-resistant Gram-negative bacteria, aiming to provide a snapshot of the situation and some information on the broader context.
-
Citations
Citations to this article as recorded by- Efficacy of new generation biosorbents for the sustainable treatment of antibiotic residues and antibiotic resistance genes from polluted waste effluent
Barkha Madhogaria, Sangeeta Banerjee, Atreyee Kundu, Prasanta Dhak
Infectious Medicine.2024; 3(1): 100092. CrossRef - Evaluation of Plant-Based Silver Nanoparticles for Antioxidant Activity and Promising Wound-Healing Applications
Maria Qubtia, Shazia Akram Ghumman, Sobia Noreen, Huma Hameed, Shazia Noureen, Rizwana Kausar, Ali Irfan, Pervaiz Akhtar Shah, Hafsa Afzal, Misbah Hameed, Mohammad Raish, Maria Rana, Ajaz Ahmad, Katarzyna Kotwica-Mojzych, Yousef A. Bin Jardan
ACS Omega.2024; 9(10): 12146. CrossRef
- Efficacy of new generation biosorbents for the sustainable treatment of antibiotic residues and antibiotic resistance genes from polluted waste effluent
- Epidemiological characteristics of carbapenemase-producing Enterobacteriaceae outbreaks in the Republic of Korea between 2017 and 2022
- Hyoseon Jeong, Junghee Hyun, Yeon-Kyeng Lee
- Osong Public Health Res Perspect. 2023;14(4):312-320. Published online August 21, 2023
- DOI: https://doi.org/10.24171/j.phrp.2023.0069
- 1,077 View
- 144 Download
- 2 Crossref
-
 Graphical Abstract
Graphical Abstract
 Abstract
Abstract
 PDF
PDF 
- Objectives
We aimed to describe the epidemiological characteristics of carbapenemase-producing Enterobacteriaceae (CPE) outbreaks in healthcare settings in the Republic of Korea between 2017 and 2022.
Methods
Under the national notifiable disease surveillance system, we obtained annual descriptive statistics regarding the isolated species, carbapenemase genotype, healthcare facility type, outbreak location and duration, and number of patients affected and recommended interventions. We used epidemiological investigation reports on CPE outbreaks reported to Korea Disease Control and Prevention Agency from June 2017 to September 2022.
Results
Among the 168 reports analyzed, Klebsiella pneumoniae (85.1%) was the most frequently reported species, while K. pneumoniae carbapenemase (KPC, 82.7%) was the most common carbapenemase genotype. Both categories increased from 2017 to 2022 (p<0.01). General hospitals had the highest proportion (54.8%), while tertiary general hospitals demonstrated a decreasing trend (p<0.01). The largest proportion of outbreaks occurred exclusively in intensive care units (ICUs, 44.0%), and the frequency of concurrent outbreaks in ICUs and general wards increased over time (p<0.01). The median outbreak duration rose from 43.5 days before the coronavirus disease 2019 (COVID-19) pandemic (2017–2019) to 79.5 days during the pandemic (2020–2022) (p=0.01), and the median number of patients associated with each outbreak increased from 5.0 to 6.0 (p=0.03). Frequently recommended interventions included employee education (38.1%), and 3 or more measures were proposed for 45.2% of outbreaks.
Conclusion
In the Republic of Korea, CPE outbreaks have been consistently dominated by K. pneumoniae and KPC. The size of these outbreaks increased during the COVID-19 pandemic. Our findings highlight the need for continuing efforts to control CPE outbreaks using a multimodal approach, while considering their epidemiology. -
Citations
Citations to this article as recorded by- Comparison of clinical outcomes of patients with serial negative surveillance cultures according to a subsequent polymerase chain reaction test for carbapenemase-producing Enterobacterales
H. Seo, S. Kim, Y.W. Lee, H.S. Oh, H-S. Kim, Y.K. Kim
Journal of Hospital Infection.2024; 146: 93. CrossRef - Identifying Contact Time Required for Secondary Transmission of Clostridioides difficile Infections by Using Real-Time Locating System
Min Hyung Kim, Jaewoong Kim, Heejin Ra, Sooyeon Jeong, Yoon Soo Park, Dongju Won, Hyukmin Lee, Heejung Kim
Emerging Infectious Diseases.2024;[Epub] CrossRef
- Comparison of clinical outcomes of patients with serial negative surveillance cultures according to a subsequent polymerase chain reaction test for carbapenemase-producing Enterobacterales
- Carbapenem resistance in critically important human pathogens isolated from companion animals: a systematic literature review
- Angie Alexandra Rincón-Real, Martha Cecilia Suárez-Alfonso
- Osong Public Health Res Perspect. 2022;13(6):407-423. Published online December 16, 2022
- DOI: https://doi.org/10.24171/j.phrp.2022.0033
- 3,030 View
- 149 Download
- 4 Web of Science
- 6 Crossref
-
 Abstract
Abstract
 PDF
PDF - This study aimed to describe the presence and geographical distribution of Gram-negativebacteria considered critical on the priority list of antibiotic-resistant pathogens publishedby the World Health Organization, including carbapenem-resistant Enterobacteriaceae,carbapenem-resistant Acinetobacter spp., and carbapenem-resistant Pseudomonas aeruginosa.A systematic review of original studies published in 5 databases between 2010 and 2021 wasconducted, including genotypically confirmed carbapenem-resistant isolates obtained fromcanines, felines, and their settings. Fifty-one articles met the search criteria. Carbapenemresistant isolates were found in domestic canines and felines, pet food, and on veterinarymedical and household surfaces. The review found that the so-called “big five”—that is, the5 major carbapenemases identified worldwide in Enterobacterales (New Delhi metallo-βlactamase, active-on-imipenem, Verona integron-encoded metallo-β-lactamase, Klebsiellapneumoniae carbapenemase, and oxacillin [OXA]-48-like)—and the 3 most importantcarbapenemases from Acinetobacter spp. (OXA-23-like, OXA-40-like, and OXA-58-like) hadbeen detected in 8 species in the Enterobacteriaceae family and 5 species of glucose nonfermenting bacilli on 5 continents. Two publications used molecular analysis to confirmcarbapenem-resistant bacteria transmission between owners and dogs. Isolating criticallyimportant human carbapenem-resistant Gram-negative bacteria from domestic canines andfelines highlights the importance of including these animal species in surveillance programsand antimicrobial resistance containment plans as part of the One Health approach.
-
Citations
Citations to this article as recorded by- First report of a blaNDM-5-carrying Escherichia coli sequence type 12 isolated from a dog with pyometra in Japan
Kazuki Harada, Tadashi Miyamoto, Michiyo Sugiyama, Tetsuo Asai
Journal of Infection and Chemotherapy.2024;[Epub] CrossRef - The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2021–2022
EFSA Journal.2024;[Epub] CrossRef - Epidemiological analysis and prevention strategies in response to a shigellosis cluster outbreak: a retrospective case series in an alternative school in the Republic of Korea, 2023
Yeongseo Ahn, Sunmi Jin, Gemma Park, Hye Young Lee, Hyungyong Lee, Eunkyung Shin, Junyoung Kim, Jaeil Yoo, Yuna Kim
Osong Public Health and Research Perspectives.2024; 15(1): 68. CrossRef - The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021
EFSA Journal.2023;[Epub] CrossRef - Resistome-based surveillance identifies ESKAPE pathogens as the predominant gram-negative organisms circulating in veterinary hospitals
Flavia Zendri, Cajsa M. Isgren, Jane Devaney, Vanessa Schmidt, Rachel Rankin, Dorina Timofte
Frontiers in Microbiology.2023;[Epub] CrossRef - Unveiling the emergence of multidrug-resistant pathogens in exotic pets from France: a comprehensive study (2017-2019)
Sandro Cardoso, Aurélie Le Loc’h, Inês Marques, Anabela Almeida, Sérgio Sousa, Maria José Saavedra, Sofia Anastácio, Eduarda Silveira
One Health & Implementation Research.2023; 3(4): 161. CrossRef
- First report of a blaNDM-5-carrying Escherichia coli sequence type 12 isolated from a dog with pyometra in Japan
- Epidemiological characteristics of carbapenem-resistant Enterobacteriaceae and carbapenem-resistant Acinetobacter baumannii in a tertiary referral hospital in Korea
- Sollan Kang, Ihn Sook Jeong
- Osong Public Health Res Perspect. 2022;13(3):221-229. Published online June 30, 2022
- DOI: https://doi.org/10.24171/j.phrp.2022.0097
- 2,448 View
- 67 Download
- 1 Web of Science
- 2 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
This study aimed to identify the epidemiological characteristics of patients with carbapenem-resistant Enterobacteriaceae and Acinetobacter baumannii (CRE/ CRAB) isolates in a tertiary referral hospital in Korea. Methods: We collected and analyzed data from 528 adults admitted to a tertiary referral hospital from August 1, 2018 to February 29, 2020. The CRE/CRAB isolates were confirmed as being present at the time of patients’ admission or acquired during hospitalization based on their medical records. The t-test, chi-square test, or Fisher exact test and stepwise multiple logistic regression were performed. Results: While the proportion of community-acquired CRE/CRAB was low (6%), 20% of CRE/ CRAB isolates were identified in patients at the time of hospitalization. The risk of CRAB isolation was positively associated with mechanical ventilator use (odds ratio [OR], 3.52; 95% confidence interval [CI], 1.96−6.33) and total parenteral nutrition use (OR, 3.64; 95% CI, 1.87−7.08). Conclusion: Over 20% of CRE/CRAB isolates in a tertiary referral hospital in Korea were found at the time of patients’ admission. Furthermore, patients with mechanical ventilation and/or total parenteral nutrition tended to acquire CRAB more frequently. Thus, active surveillance for CRE/CRAB at the time of hospitalization is strongly required, particularly for patients who are expected to require mechanical ventilation or total parenteral nutrition. -
Citations
Citations to this article as recorded by- Epidemiological analysis and prevention strategies in response to a shigellosis cluster outbreak: a retrospective case series in an alternative school in the Republic of Korea, 2023
Yeongseo Ahn, Sunmi Jin, Gemma Park, Hye Young Lee, Hyungyong Lee, Eunkyung Shin, Junyoung Kim, Jaeil Yoo, Yuna Kim
Osong Public Health and Research Perspectives.2024; 15(1): 68. CrossRef - Epidemiology and Risk Factors of Carbapenemase-Producing Enterobacteriaceae Acquisition and Colonization at a Korean Hospital over 1 Year
Hye-Jin Kim, Jung-Hee Hyun, Hyo-Seon Jeong, Yeon-Kyeng Lee
Antibiotics.2023; 12(4): 759. CrossRef
- Epidemiological analysis and prevention strategies in response to a shigellosis cluster outbreak: a retrospective case series in an alternative school in the Republic of Korea, 2023
- Yersinia pestis antibiotic resistance: a systematic review
- Chen Lei, Suresh Kumar
- Osong Public Health Res Perspect. 2022;13(1):24-36. Published online February 18, 2022
- DOI: https://doi.org/10.24171/j.phrp.2021.0288
- 7,213 View
- 248 Download
- 8 Web of Science
- 10 Crossref
-
 Abstract
Abstract
 PDF
PDF - Yersinia pestis, the cause of plague and a potential biological weapon, has always been a threatening pathogen. Some strains of Y. pestis have varying degrees of antibiotic resistance. Thus, this systematic review was conducted to alert clinicians to this pathogen’s potential antimicrobial resistance. A review of the literature was conducted for experimental reports and systematic reviews on the topics of plague, Y. pestis, and antibiotic resistance. From 1995 to 2021, 7 Y. pestis isolates with 4 antibiotic resistance mechanisms were reported. In Y. pestis 17/95, 16/95, and 2180H, resistance was mediated by transferable plasmids. Each plasmid contained resistance genes encoded within specific transposons. Strain 17/95 presented multiple drug resistance, since plasmid 1202 contained 10 resistance determinants. Strains 16/95 and 2180H showed single antibiotic resistance because both additional plasmids in these strains carried only 1 antimicrobial determinant. Strains 12/87, S19960127, 56/13, and 59/13 exhibited streptomycin resistance due to an rpsl gene mutation, a novel mechanism that was discovered recently. Y. pestis can acquire antibiotic resistance in nature not only via conjugative transfer of antimicrobial-resistant plasmids from other bacteria, but also by gene point mutations. Global surveillance should be strengthened to identify antibiotic-resistant Y. pestis strains by whole-genome sequencing and drug susceptibility testing.
-
Citations
Citations to this article as recorded by- Seek and you shall find: Yersinia enterocolitica in Ireland’s drinking water
James Powell, Maureen Daly, Nuala H. O’Connell, Colum P. Dunne
Irish Journal of Medical Science (1971 -).2024;[Epub] CrossRef - A novel sORF gene mutant strain of Yersinia pestis vaccine EV76 offers enhanced safety and improved protection against plague
Xiao Guo, Youquan Xin, Zehui Tong, Shiyang Cao, Yuan Zhang, Gengshan Wu, Hongyan Chen, Tong Wang, Yajun Song, Qingwen Zhang, Ruifu Yang, Zongmin Du, Gregory P. Priebe
PLOS Pathogens.2024; 20(3): e1012129. CrossRef - Rapid Induction of Protective Immunity against Pneumonic Plague by Yersinia pestis Polymeric F1 and LcrV Antigens
Moshe Aftalion, Avital Tidhar, Yaron Vagima, David Gur, Ayelet Zauberman, Tzvi Holtzman, Arik Makovitzki, Theodor Chitlaru, Emanuelle Mamroud, Yinon Levy
Vaccines.2023; 11(3): 581. CrossRef - Antibiotic resistance in Neisseria gonorrhoeae: broad-spectrum drug target identification using subtractive genomics
Umairah Natasya Mohd Omeershffudin, Suresh Kumar
Genomics & Informatics.2023; 21(1): e5. CrossRef - Polyclonal Antibodies Derived from Transchromosomic Bovines Vaccinated with the Recombinant F1-V Vaccine Increase Bacterial Opsonization In Vitro and Protect Mice from Pneumonic Plague
Sergei S. Biryukov, Hua Wu, Jennifer L. Dankmeyer, Nathaniel O. Rill, Christopher P. Klimko, Kristi A. Egland, Jennifer L. Shoe, Melissa Hunter, David P. Fetterer, Ju Qiu, Michael L. Davies, Christoph L. Bausch, Eddie J. Sullivan, Thomas Luke, Christopher
Antibodies.2023; 12(2): 33. CrossRef - New Bacteriophages with Podoviridal Morphotypes Active against Yersinia pestis: Characterization and Application Potential
Tamar Suladze, Ekaterine Jaiani, Marina Darsavelidze, Maia Elizbarashvili, Olivier Gorge, Ia Kusradze, Tamar Kokashvili, Nino Lashkhi, George Tsertsvadze, Nino Janelidze, Svetlana Chubinidze, Marina Grdzelidze, Shota Tsanava, Eric Valade, Marina Tediashvi
Viruses.2023; 15(7): 1484. CrossRef -
Characterization of Mu-Like
Yersinia
Phages Exhibiting Temperature Dependent Infection
Biao Meng, Zhizhen Qi, Xiang Li, Hong Peng, Shanzheng Bi, Xiao Wei, Yan Li, Qi Zhang, Xiaoqing Xu, Haihong Zhao, Xiaoyan Yang, Changjun Wang, Xiangna Zhao, Olaya Rendueles
Microbiology Spectrum.2023;[Epub] CrossRef -
Ancient
Yersinia pestis
genomes lack the virulence-associated Ypf
Φ
prophage present in modern pandemic strains
Joanna H. Bonczarowska, Julian Susat, Ben Krause-Kyora, Dorthe Dangvard Pedersen, Jesper Boldsen, Lars Agersnap Larsen, Lone Seeberg, Almut Nebel, Daniel Unterweger
Proceedings of the Royal Society B: Biological Sci.2023;[Epub] CrossRef - A situation analysis of the current plague outbreak in the Demographic Republic of Congo and counteracting strategies – Correspondence
Ranjit Sah, Abdullah Reda, Rachana Mehta, Ranjan K. Mohapatra, Kuldeep Dhama
International Journal of Surgery.2022; 105: 106885. CrossRef - Antimicrobial resistance in Klebsiella pneumoniae: identification of bacterial DNA adenine methyltransferase as a novel drug target from hypothetical proteins using subtractive genomics
Umairah Natasya Mohd Omeershffudin, Suresh Kumar
Genomics & Informatics.2022; 20(4): e47. CrossRef
- Seek and you shall find: Yersinia enterocolitica in Ireland’s drinking water
- Prevalence of plasmid-mediated AmpC β-lactamases among uropathogenic Escherichia coli isolates in southwestern Iran
- Nabi Jomehzadeh, Khadijeh Ahmadi, Zahra Rahmani
- Osong Public Health Res Perspect. 2021;12(6):390-395. Published online December 1, 2021
- DOI: https://doi.org/10.24171/j.phrp.2021.0272
- 4,961 View
- 92 Download
- 5 Web of Science
- 6 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
This study was undertaken to evaluate AmpC β-lactamase-producing Escherichia coli urine isolates and to characterize the frequency of plasmid-mediated AmpC (pAmpC)-encoding genes.
Methods
Antimicrobial susceptibility tests were performed using the disk diffusion technique. AmpC β-lactamase production was assessed with a phenotypic inhibitor-based method. The presence of 6 pAmpC-encoding cluster genes was detected by multiplex polymerase chain reaction (PCR).
Results
The proportion of antibiotic resistance of E. coli isolates ranged from 7.4% to 90.5%, and more than half (51.6%) of the total isolates were multidrug-resistant (MDR). Among the 95 E. coli isolates, 60 (63.2%) were found to be cefoxitin-resistant, but only 14 (14.7%) isolates were confirmed as AmpC β-lactamase-producers. In the PCR assay, pAmpC-encoding genes were found in 15 (15.8%) isolates, and blaDHA was the most prevalent type. However, blaFOX, blaMOX, and blaACC genes were not detected in the isolates.
Conclusion
Our findings contributed valuable information concerning antibiotic resistance, confirmatory phenotypic testing for AmpC production, and pAmpC β-lactamase gene content in E. coli isolates in southwestern Iran. The level of MDR recorded in AmpC-producing strains of this study was worrying; therefore, implementing strong infection control approaches to reduce the MDR burden is recommended. -
Citations
Citations to this article as recorded by- A review of the mechanisms that confer antibiotic resistance in pathotypes of E. coli
Sina Nasrollahian, Jay P. Graham, Mehrdad Halaji
Frontiers in Cellular and Infection Microbiology.2024;[Epub] CrossRef - Association Between Uropathogenic Escherichia coli Virulence Genes and Severity of Infection and Resistance to Antibiotics
Sofía Alejandra Fonseca-Martínez, Ruth Aralí Martínez-Vega, Ana Elvira Farfán-García, Clara Isabel González Rugeles, Libeth Yajaira Criado-Guerrero
Infection and Drug Resistance.2023; Volume 16: 3707. CrossRef - Extended-spectrum β-lactamases producing Enterobacteriaceae (ESBL-PE) prevalence in Nepal: A systematic review and meta-analysis
Christina Khadka, Manita Shyaula, Gopiram Syangtan, Shrijana Bista, Reshma Tuladhar, Anjana Singh, Dev Raj Joshi, Lok R. Pokhrel, Prabin Dawadi
Science of The Total Environment.2023; 901: 166164. CrossRef - Prevalence of Extended-Spectrum β-Lactamase-Resistant Genes in Escherichia coli Isolates from Central China during 2016–2019
Zui Wang, Qin Lu, Xiaohui Mao, Li Li, Junfeng Dou, Qigai He, Huabin Shao, Qingping Luo
Animals.2022; 12(22): 3191. CrossRef - Molecular detection and characterization of Shigella spp. harboring extended-spectrum β-lactamase genes in children with diarrhea in northwest Iran
Sahar Sabour, Amir Teimourpour, Jafar Mohammadshahi, Hadi Peeridogaheh, Roghayeh Teimourpour, Taher Azimi, Zahra Hosseinali
Molecular and Cellular Pediatrics.2022;[Epub] CrossRef - Plasmid-mediated AmpC β-Lactamase gene analysis in Klebsiella Pneumoniae clinical isolates
Nabi Jomehzadeh, Khadijeh Ahmadi, Hasti Shaabaninejad, Gholamali Eslami
Biomedical and Biotechnology Research Journal (BBR.2022; 6(4): 582. CrossRef
- A review of the mechanisms that confer antibiotic resistance in pathotypes of E. coli
- Characterization of Antimicrobial Susceptibility, Extended-Spectrum β-Lactamase Genes and Phylogenetic Groups of Enteropathogenic
Escherichia coli Isolated from Patients with Diarrhea - Erfaneh Jafari, Saeid Mostaan, Saeid Bouzari
- Osong Public Health Res Perspect. 2020;11(5):327-333. Published online October 22, 2020
- DOI: https://doi.org/10.24171/j.phrp.2020.11.5.09
- 7,708 View
- 104 Download
- 5 Web of Science
- 7 Crossref
-
 Abstract
Abstract
 PDF
PDF Objectives Infectious diarrhea is one of the most common causes of pediatric death worldwide and enteropathogenic
Escherichia coli (EPEC) is one of the main causes. There are 2 subgroups of EPEC, typical and atypical, based on the presence or absence of bundle forming pili (bfp), of which atypical EPEC is considered less virulent, but not less pathogenic. Antimicrobial resistance towards atypical EPEC among children is growing and is considered a major problem. In this study the pattern of antibiotic resistance in clinical isolates was determined.Methods Using 130 isolates, antibiotic resistance patterns and phenotypes were assessed, and genotypic profiles of extended spectrum β-lactamase (ESBL) production using disc diffusion and PCR was carried out. Phylogenetic groups were analyzed using quadruplex PCR.
Results There were 65
E. coli isolates identified as atypical EPEC by PCR, among which the highest antibiotic resistance was towards ampicillin, followed by trimethoprim-sulfamethoxazole, and tetracycline. Multidrug resistance was detected in 44.6% of atypical EPEC isolates. Around 33% of isolates were determined to be extended spectrum β-lactamase producers, and in 90% of isolates, genes responsible for ESBL production could be detected. Moreover, the majority of atypical EPEC strains belonged to Group E, followed by Groups B1, B2 and C.Conclusion High rates of multidrug resistance and ESBL production among atypical EPEC isolates warrant periodical surveillance studies to select effective antibiotic treatment for patients. It is considered a critical step to manage antibiotic resistance by avoiding unnecessary prescriptions for antibiotics.
-
Citations
Citations to this article as recorded by- Phenotypic and molecular characterization of enteropathogenic Escherichia coli and Salmonella spp. causing childhood diarrhoea in Awka, South-Eastern Nigeria
Ifeanyi Emmanuel Nwike, Malachy Chigozie Ugwu, Peter Chika Ejikeugwu, Nonye Treasure Ujam, Ifeanyichukwu Romanus Iroha, Charles Okechukwu Esimone
Bulletin of the National Research Centre.2023;[Epub] CrossRef - Antimicrobial Resistance and Biofilm Formation of Escherichia coli Isolated from Pig Farms and Surroundings in Bulgaria
Mila D. Kaleva, Yana Ilieva, Maya Margaritova Zaharieva, Lyudmila Dimitrova, Tanya Chan Kim, Iva Tsvetkova, Yordan Georgiev, Petya Orozova, Krasimir Nedev, Hristo Najdenski
Microorganisms.2023; 11(8): 1909. CrossRef - Isolation and Molecular Characterization of Proteus mirabilis Bacteria from Poultry Meat in the Iraqi Market and a Study of their Resistance to some Antibiotics
Ammar H. Hamed, Ahmed I. Alnazzal
IOP Conference Series: Earth and Environmental Sci.2023; 1262(6): 062019. CrossRef - Virulence and phylogenetic analysis of enteric pathogenic Escherichia coli isolated from children with diarrhoea in South Africa
Ntando W. Alfinete, John Y. Bolukaoto, Lee Heine, Natasha Potgieter, Tobias G. Barnard
International Journal of Infectious Diseases.2022; 114: 226. CrossRef - Prevalence of extended-spectrum β-lactamases, AmpC, and carbapenemases in Proteus mirabilis clinical isolates
Mona Shaaban, Soha Lotfy Elshaer, Ola A. Abd El-Rahman
BMC Microbiology.2022;[Epub] CrossRef - Enteropathogenic Escherichia coli—A Summary of the Literature
Anca Delia Mare, Cristina Nicoleta Ciurea, Adrian Man, Bianca Tudor, Valeriu Moldovan, Luminița Decean, Felicia Toma
Gastroenterology Insights.2021; 12(1): 28. CrossRef - Prevalence of Antibiotic-Resistant Escherichia coli Isolated from Swine Faeces and Lagoons in Bulgaria
Lyudmila Dimitrova, Mila Kaleva, Maya M. Zaharieva, Christina Stoykova, Iva Tsvetkova, Maya Angelovska, Yana Ilieva, Vesselin Kussovski, Sevda Naydenska, Hristo Najdenski
Antibiotics.2021; 10(8): 940. CrossRef
- Phenotypic and molecular characterization of enteropathogenic Escherichia coli and Salmonella spp. causing childhood diarrhoea in Awka, South-Eastern Nigeria
- Analysis of Resistance to Macrolide–Lincosamide–Streptogramin B Among
mecA -PositiveStaphylococcus Aureus Isolates - Mahmoud Khodabandeh, Mohsen Mohammadi, Mohammad Reza Abdolsalehi, Azadeh Alvandimanesh, Mehrdad Gholami, Meysam Hasannejad Bibalan, Abazar Pournajaf, Ramin Kafshgari, Ramazan Rajabnia
- Osong Public Health Res Perspect. 2019;10(1):25-31. Published online February 28, 2019
- DOI: https://doi.org/10.24171/j.phrp.2019.10.1.06
- 6,087 View
- 186 Download
- 27 Crossref
-
 Abstract
Abstract
 PDF
PDF Objectives Genetic determinants conferring resistance to macrolide, lincosamide, and streptogramin B (MLSB) via ribosomal modification such as,
erm ,msrA/B andereA/B genes are distributed in bacteria. The main goals of this work were to evaluate the dissemination of MLSB resistance phenotypes and genotypes in methicillin-resistantStaphylococcus aureus (MRSA) isolates collected from clinical samples.Methods A total of 106 MRSA isolates were studied. Isolates were recovered from 3 hospitals in Tehran between May 2016 to July 2017. The prevalence of MLSB-resistant strains were determined by D-test, and then M-PCR was performed to identify genes encoding resistance to macrolides, lincosamides, and streptogramins in the tested isolates.
Results The frequency of constitutive resistance MLSB, inducible resistance MLSB and MSB resistance were 56.2%, 22.9%, and 16.6%, respectively. Of 11 isolates with the inducible resistance MLSB phenotype,
ermC ,ermB ,ermA andereA were positive in 81.8%, 63.6%, 54.5% and 18.2% of these isolates, respectively. In isolates with the constitutive resistance MLSB phenotype, the prevalence ofermA ,ermB ,ermC ,msrA ,msrB ,ereA andereB were 25.9%, 18.5%, 44.4%, 0.0%, 0.0%, 11.1% and 0.0%, respectively.Conclusion Clindamycin is commonly administered in severe MRSA infections depending upon the antimicrobial susceptibility findings. This study showed that the D-test should be used as an obligatory method in routine disk diffusion assay to detect inducible clindamycin resistance in MRSA so that effective antibiotic treatment can be provided.
-
Citations
Citations to this article as recorded by- Inducible Clindamycin Resistance in Staphylococcus aureus Isolates in Kermanshah, Iran
Zahra Jahanbakhshi, Jamileh Nowroozi, Zahra Kahrarian, Azin Tariniya Gilani, Mohadeseh Ahmadvand, Nasrollah Sohrabi
Journal of Clinical Research in Paramedical Scienc.2024;[Epub] CrossRef - Antimicrobial Resistance and the Prevalence of the Panton-Valentine Leukocidin Gene among Clinical Isolates of Staphylococcus aureus in Lithuania
Agnė Kirkliauskienė, Jonas Kriščiūnas, Jolanta Miciulevičienė, Daiva Radzišauskienė, Tomas Kačergius, Maksim Bratchikov, Lina Kaplerienė
Polish Journal of Microbiology.2024; 73(1): 21. CrossRef - Characterization of oxacillin-resistant Staphylococcus lugdunensis isolated from sterile body fluids in a medical center in Taiwan: A 12-year longitudinal epidemiological study
Shih-Cheng Chang, Jazon Harl Hidrosollo, Lee-Chung Lin, Yu-Hsiang Ou, Cheng-Yen Kao, Jang-Jih Lu
Journal of Microbiology, Immunology and Infection.2023; 56(2): 292. CrossRef - A new insight into the potential drivers of antibiotic resistance gene enrichment in the collembolan gut association with antibiotic and non-antibiotic agents
Yi-Fei Wang, Tian-Gui Cai, Zhe-Lun Liu, Hui-Ling Cui, Dong Zhu, Min Qiao
Journal of Hazardous Materials.2023; 451: 131133. CrossRef - Phenotypic and Genotypic Characterization of Macrolide-Lincosamide-Streptogramin Resistance in Staphylococcus aureus Isolates from Bovine and Human
Ozgul Gulaydin, Kemal Gurturk, Ismail Hakki Ekin, Ziya Ilhan, Cigdem Arabaci
Acta Veterinaria.2023; 73(1): 102. CrossRef - Encapsulated peracetic acid as a valid broad-spectrum antimicrobial alternative, leading to beneficial microbiota compositional changes and enhanced performance in broiler chickens
Salvatore Galgano, Leah Conway, Nikki Dalby, Adrian Fellows, Jos G. M. Houdijk
Journal of Animal Science and Biotechnology.2023;[Epub] CrossRef - Metagenomic investigation reveals bacteriophage-mediated horizontal transfer of antibiotic resistance genes in microbial communities of an organic agricultural ecosystem
Yujie Zhang, Ai Kitazumi, Yen-Te Liao, Benildo G. de los Reyes, Vivian C. H. Wu, Thomas G. Denes
Microbiology Spectrum.2023;[Epub] CrossRef - New update on molecular diversity of clinical Staphylococcus aureus isolates in Iran: antimicrobial resistance, adhesion and virulence factors, biofilm formation and SCCmec typing
Mahtab Tabandeh, Hami Kaboosi, Mojtaba Taghizadeh Armaki, Abazar Pournajaf, Fatemeh Peyravii Ghadikolaii
Molecular Biology Reports.2022; 49(4): 3099. CrossRef - Inducible Clindamycin-Resistant Staphylococcus aureus Strains in Africa: A Systematic Review
Muluneh Assefa, Faham Khamesipour
International Journal of Microbiology.2022; 2022: 1. CrossRef - Genetic Diversity and Virulence Profile of Methicillin and Inducible Clindamycin-Resistant Staphylococcus aureus Isolates in Western Algeria
Zahoua Mentfakh Laceb, Seydina M. Diene, Rym Lalaoui, Mabrouk Kihal, Fella Hamaidi Chergui, Jean-Marc Rolain, Linda Hadjadj
Antibiotics.2022; 11(7): 971. CrossRef - Characteristics of antibiotic resistance gene distribution in rainfall runoff and combined sewer overflow
Xin-rong Pan, Lei Chen, Li-ping Zhang, Jian-e Zuo
Environmental Science and Pollution Research.2022; 30(11): 30766. CrossRef - Staphylococcus aureus from Subclinical Cases of Mastitis in Dairy Cattle in Poland, What Are They Hiding? Antibiotic Resistance and Virulence Profile
Edyta Kaczorek-Łukowska, Joanna Małaczewska, Patrycja Sowińska, Marta Szymańska, Ewelina Agnieszka Wójcik, Andrzej Krzysztof Siwicki
Pathogens.2022; 11(12): 1404. CrossRef - Surveillance of osteoarticular infections caused by Staphylococcus aureus in a paediatric hospital in Mexico City
Nancy Evelyn Aguilar-Gómez, Jocelin Merida-Vieyra, Oscar Daniel Isunza-Alonso, María Gabriela Morales-Pirela, Oscar Colín-Martínez, Enrique Josué Juárez-Benítez, Silvestre García de la Puente, Alejandra Aquino-Andrade
Frontiers in Cellular and Infection Microbiology.2022;[Epub] CrossRef - Molecular typing, biofilm formation, and analysis of adhesion factors in Staphylococcus aureus strains isolated from urinary tract infections
Masoumeh Navidinia, Anis Mohammadi, Reza Arjmand, Masoud Dadashi, Mehdi Goudarzi
Gene Reports.2021; 22: 101008. CrossRef - Molecular characterization of invasive Staphylococcus aureus strains isolated from patients with diabetes in Iran: USA300 emerges as the major type
Zahra Tayebi, Maryam Fazeli, Ali Hashemi, Saeed Abdi, Masoud Dadashi, Mohammad Javad Nasiri, Mehdi Goudarzi
Infection, Genetics and Evolution.2021; 87: 104679. CrossRef - Predominance of PVL-negative community-associated methicillin-resistant Staphylococcus aureus sequence type 8 in newly diagnosed HIV-infected adults, Tanzania
Joel Manyahi, Sabrina J. Moyo, Said Aboud, Nina Langeland, Bjørn Blomberg
European Journal of Clinical Microbiology & Infect.2021; 40(7): 1477. CrossRef - Molecular characterization of Staphylococcus aureus strains isolated from hospitalized patients based on coagulase gene polymorphism analysis: High frequency of vancomycin-intermediate S. aureus and the emergence of coagulase type II in Iran
Malihe Soltani, Bahareh Hajikhani, Samira Zamani, Mehrdad Haghighi, Ali Hashemi, Mohammad Javad Nasiri, Masoud Dadashi, Behzad Pourhossein, Mehdi Goudarzi
Gene Reports.2021; 23: 101078. CrossRef - Inducible clindamycin resistance among clinical Staphylococcus aureus strains in Iran: A contemporaneous systematic review and meta-analysis
Mojtaba Memariani, Hamed Memariani, Hamideh Moravvej
Gene Reports.2021; 23: 101104. CrossRef - Association of Macrolide Resistance Genotypes and Synergistic Antibiotic Combinations for Combating Macrolide-Resistant MRSA Recovered from Hospitalized Patients
Amr S. Bishr, Salma M. Abdelaziz, Ibrahim S. Yahia, Mahmoud A. Yassien, Nadia A. Hassouna, Khaled M. Aboshanab
Biology.2021; 10(7): 624. CrossRef - Relationship between MLSB resistance and the prevalent virulence genotypes among Bulgarian Staphylococcus aureus isolates
Virna-Maria Tsitou, Ivan Mitov, Raina Gergova
Acta Microbiologica et Immunologica Hungarica.2021; 68(1): 55. CrossRef - Occurrence, Antibiotic Resistance, Virulence Factors, and Genetic Diversity of Bacillus spp. from Public Hospital Environments in South Africa
Zamile N. Mbhele, Christiana O. Shobo, Daniel G. Amoako, Oliver T. Zishiri, Linda A. Bester
Microbial Drug Resistance.2021; 27(12): 1692. CrossRef - Bacterial Antibiotic Resistance: The Most Critical Pathogens
Giuseppe Mancuso, Angelina Midiri, Elisabetta Gerace, Carmelo Biondo
Pathogens.2021; 10(10): 1310. CrossRef - Mechanisms of Resistance to Macrolide Antibiotics among Staphylococcus aureus
Maria Miklasińska-Majdanik
Antibiotics.2021; 10(11): 1406. CrossRef - Genetic Characterization of Methicillin-Resistant Staphylococcus aureus Isolates from Human Bloodstream Infections: Detection of MLSB Resistance
Vanessa Silva, Sara Hermenegildo, Catarina Ferreira, Célia M. Manaia, Rosa Capita, Carlos Alonso-Calleja, Isabel Carvalho, José Eduardo Pereira, Luis Maltez, José L. Capelo, Gilberto Igrejas, Patrícia Poeta
Antibiotics.2020; 9(7): 375. CrossRef - Genetic analysis of methicillin‐susceptible Staphylococcus aureus clinical isolates: High prevalence of multidrug‐resistant ST239 with strong biofilm‐production ability
Hossein Goudarzi, Mehdi Goudarzi, Fattaneh Sabzehali, Maryam Fazeli, Alireza Salimi Chirani
Journal of Clinical Laboratory Analysis.2020;[Epub] CrossRef - Emergence and spread of coagulase type III and staphylococcal cassette chromosome mec type IV among mupirocin-resistant Staphylococcus aureus isolated from wound infections
Mirmohammad Miri, Maryam Fazeli, Anahita Amirpour, Mohammad Javad Nasiri, Ramin Pouriran, Mehdi Goudarzi
Gene Reports.2020; 21: 100858. CrossRef - Resistance profile to antimicrobials agents in methicillin-resistant Staphylococcus aureus isolated from hospitals in South Brazil between 2014-2019
Adriana Medianeira Rossato, Muriel Primon-Barros, Lisiane da Luz Rocha, Keli Cristine Reiter, Cícero Armídio Gomes Dias, Pedro Alves d’Azevedo
Revista da Sociedade Brasileira de Medicina Tropic.2020;[Epub] CrossRef
- Inducible Clindamycin Resistance in Staphylococcus aureus Isolates in Kermanshah, Iran
- High Prevalence of Class 1 to 3 Integrons Among Multidrug-Resistant Diarrheagenic
Escherichia coli in Southwest of Iran - Mohammad Kargar, Zahra Mohammadalipour, Abbas Doosti, Shahrokh Lorzadeh, Alireza Japoni-Nejad
- Osong Public Health Res Perspect. 2014;5(4):193-198. Published online August 31, 2014
- DOI: https://doi.org/10.1016/j.phrp.2014.06.003
- 3,195 View
- 24 Download
- 44 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
Horizontal transfer of integrons is one of the important factors that can contribute to the occurrence of multidrug-resistant (MDR) bacteria. This study aimed to determine the prevalence of integrons among MDR Escherichia coli strains isolated from stool specimens and investigate the associations between the existence of integrons and MDR properties in the southwest of Iran.
Methods
There were 164 E. coli strains isolated from January 2012 to June 2012. Fecal specimens identified as E. coli by the conventional methods. Subsequently the antibiotic resistance was assessed using Clinical and Laboratory Standard Institute criteria. The presence of class 1–3 integrons and embedded gene cassettes was verified using specific primers by multiplex polymerase chain reaction assay.
Results
Among a total of 164 studied samples, 69 (42.07%) isolates were multidrug resistant. Class 1 and class 2 integrons were present in 78.26% and 76.81% MDR isolates, respectively. For the first time in Iran, class 3 integron was observed in 26.09% MDR isolates. Significant correlations were identified between: class 1 integron and resistance to amikacin, gentamicin, chloramphenicol, ampicillin, tetracycline, nalidixic acid, and co-trimoxazole; class 2 integron and resistance to aminoglycosides, co-trimoxazole, cefalexin, ampicillin, and chloramphenicol; and class 3 integron and resistance to gentamicin, kanamycin, and streptomycin.
Conclusion
Our results indicate that integrons are common among MDR isolates and they can be used as a marker for the identification of MDR isolates. Therefore, due to the possibility of a widespread outbreak of MDR isolates, molecular surveillance and sequencing of the integrons in other parts of the country is recommended. -
Citations
Citations to this article as recorded by- Municipal wastewater treatment plant showing a potential reservoir for clinically relevant MDR bacterial strains co-occurrence of ESBL genes and integron-integrase genes
Kuldeep Soni, David Kothamasi, Ram Chandra
Journal of Environmental Management.2024; 351: 119938. CrossRef - Detection of virulence factor genes, antibiotic resistance genes and biofilm formation in clinical Gram-negative bacteria and first report from Türkiye of K.oxytoca carrying both blaOXA-23 and blaOXA-51 genes
Azer Özad Düzgün, Gamze Yüksel
Biologia.2023; 78(8): 2245. CrossRef - Antimicrobial Resistance in Romania: Updates on Gram-Negative ESCAPE Pathogens in the Clinical, Veterinary, and Aquatic Sectors
Ilda Czobor Barbu, Irina Gheorghe-Barbu, Georgiana Alexandra Grigore, Corneliu Ovidiu Vrancianu, Mariana Carmen Chifiriuc
International Journal of Molecular Sciences.2023; 24(9): 7892. CrossRef - Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolated from Food-Producing Animals in Tamaulipas, Mexico
Antonio Mandujano, Diana Verónica Cortés-Espinosa, José Vásquez-Villanueva, Paulina Guel, Gildardo Rivera, Karina Juárez-Rendón, Wendy Lizeth Cruz-Pulido, Guadalupe Aguilera-Arreola, Abraham Guerrero, Virgilio Bocanegra-García, Ana Verónica Martínez-Vázqu
Antibiotics.2023; 12(6): 1010. CrossRef - Antimicrobial susceptibility and integrons detection among extended-spectrum β-lactamase producing Enterobacteriaceae isolates in patients with urinary tract infection
Karzan Taha Abubaker, Khanda Abdulateef Anwar
PeerJ.2023; 11: e15429. CrossRef - Brucella abortus antigen omp25 vaccines: Development and targeting based on Lactococcus lactis
Somaye Tirbakhsh Gouran, Abbas Doosti, Mohammad Saeid Jami
Veterinary Medicine and Science.2023; 9(4): 1908. CrossRef - Integrons in the development of antimicrobial resistance: critical review and perspectives
Basharat Ahmad Bhat, Rakeeb Ahmad Mir, Hafsa Qadri, Rohan Dhiman, Abdullah Almilaibary, Mustfa Alkhanani, Manzoor Ahmad Mir
Frontiers in Microbiology.2023;[Epub] CrossRef - Tracing the Evolutionary Pathways of Serogroup O78 Avian Pathogenic Escherichia coli
Eun-Jin Ha, Seung-Min Hong, Seung-Ji Kim, Sun-Min Ahn, Ho-Won Kim, Kang-Seuk Choi, Hyuk-Joon Kwon
Antibiotics.2023; 12(12): 1714. CrossRef - Molecular Detection of Integrons, Colistin and β-lactamase Resistant Genes in Salmonella enterica Serovars Enteritidis and Typhimurium Isolated from Chickens and Rats Inhabiting Poultry Farms
Tsepo Ramatla, Kealeboga Mileng, Rendani Ndou, Nthabiseng Mphuti, Michelo Syakalima, Kgaugelo E. Lekota, Oriel M.M. Thekisoe
Microorganisms.2022; 10(2): 313. CrossRef - Prevalence of the Integrons and ESBL Genes in Multidrug-Resistant Strains of Escherichia coli Isolated from Urinary Tract Infections, Ardabil, Iran
Soheyla Barzegar, Mohsen Arzanlou, Amir Teimourpour, Majid Esmaelizad, Mehdi Yousefipour, Jafar MohammadShahi, Roghayeh Teimourpour
Iranian Journal of Medical Microbiology.2022; 16(1): 56. CrossRef - Migration of antibiotic resistance genes and evolution of flora structure in the Xenopus tropicalis intestinal tract with combined exposure to roxithromycin and oxytetracycline
Xiaojun Lin, Yanbin Xu, Ruiqi Han, Wenshi Luo, Li Zheng
Science of The Total Environment.2022; 820: 153176. CrossRef - Investigation of class 1 integrons and biofilm formation in multi-drug resistance uropathogenic Escherichia coli isolated from patients with urinary tract infection in shohadaye qom hospital, Iran
Ahmad Khorshidi, NadiaMohammad Zadeh, Azad Khaledi, GholamAbbas Moosavi, Ali Shakerimoghaddam, Azade Matinpur
International Archives of Health Sciences.2022; 9(1): 47. CrossRef - Prevalence and characterisation of antimicrobial resistance genes and class 1 and 2 integrons in multiresistant Escherichia coli isolated from poultry production
Przemysław Racewicz, Michał Majewski, Hanna Biesiada, Sebastian Nowaczewski, Jarosław Wilczyński, Danuta Wystalska, Magdalena Kubiak, Marcin Pszczoła, Zofia E. Madeja
Scientific Reports.2022;[Epub] CrossRef - Prevalence of integrons in multidrug-resistant Escherichia coli isolates from waters and vegetables in Nsukka and Enugu, Southeast Nigeria
Chinyere B. Chigor, Ini-Abasi I. Ibangha, Nkechinyere O. Nweze, Valentino C. Onuora, Chizoba A. Ozochi, Yinka Titilawo, Matthew C. Enebe, Tatyana N. Chernikova, Peter N. Golyshin, Vincent N. Chigor
Environmental Science and Pollution Research.2022; 29(40): 60945. CrossRef - Antimicrobial resistance and genetic diversity of Staphylococcus aureus collected from livestock, poultry and humans
Sangeeta Rao, Lyndsey Linke, Roberta Magnuson, Linzy Jauch, Doreene R. Hyatt
One Health.2022; 15: 100407. CrossRef - The Emergence of Carbapenem-Resistant Gram-Negative Bacteria in Mizoram, Northeast India
Vanlalruati S. C. Ralte, Archana Loganathan, Prasanth Manohar, Christine Vanlalbiakdiki Sailo, Zothan Sanga, Lalremruata Ralte, John Zothanzama, Sebastian Leptihn, Ramesh Nachimuthu, Nachimuthu Senthil Kumar
Microbiology Research.2022; 13(3): 342. CrossRef - Common Etiological Agents in Adult Patients with Gastroenteritis from Central Iran
Elnaz Abbasi, Alex van Belkum, Ehsanollah Ghaznavi-Rad
Microbial Drug Resistance.2022; 28(11): 1043. CrossRef - Antibiotic Resistance in Proteus mirabilis: Mechanism, Status, and Public Health Significance
Ebtehal Alqurashi, Khaled Elbanna, Iqbal Ahmad, Hussein H. Abulreesh
Journal of Pure and Applied Microbiology.2022; 16(3): 1550. CrossRef - Antibiotic Susceptibility Profiles and Resistance Mechanisms to β-Lactams and Polymyxins of Escherichia coli from Broilers Raised under Intensive and Extensive Production Systems
Mariana Ferreira, Célia Leão, Lurdes Clemente, Teresa Albuquerque, Ana Amaro
Microorganisms.2022; 10(10): 2044. CrossRef - Antimicrobial Resistance, Integron Carriage, and Fluoroquinolone Resistance Genes in Acinetobacte baumannii Isolates
Parastoo Ashouri, Jafar Mohammadshahi, Vajihe Sadat Nikbin, Hadi Peeridogaheh, Behnam Mohammadi-Ghalehbin, Soheila Refahi, Amir Teimourpour, Majid Esmaelizad, Hafez Mirzaneghad, Roghayeh Teimourpour
Archives of Clinical Infectious Diseases.2022;[Epub] CrossRef - Multidrug resistance-encoding gene in Citrobacter freundii isolated from healthy laying chicken in Blitar District, Indonesia
Adiana Mutamsari Witaningrum, Freshinta Jellia Wibisono, Dian Ayu Permatasari, Mustofa Helmi Effendi, Emmanuel Nnabuike Ugbo
International Journal of One Health.2022; : 161. CrossRef - Evaluation of Retail Meat as a Source of ESBL Escherichia coli in Tamaulipas, Mexico
Ana Verónica Martínez-Vázquez, Antonio Mandujano, Eduardo Cruz-Gonzalez, Abraham Guerrero, Jose Vazquez, Wendy Lizeth Cruz-Pulido, Gildardo Rivera, Virgilio Bocanegra-García
Antibiotics.2022; 11(12): 1795. CrossRef - Characterization of Integrons and Quinolone Resistance in Clinical Escherichia coli Isolates in Mansoura City, Egypt
Shaymaa H. Abdel-Rhman, Rehab M. Elbargisy, Dina E. Rizk, Ahmed Majeed Al-Shammari
International Journal of Microbiology.2021; 2021: 1. CrossRef - Prevalence and Characterization of ESBL/AmpC Producing Escherichia coli from Fresh Meat in Portugal
Lurdes Clemente, Célia Leão, Laura Moura, Teresa Albuquerque, Ana Amaro
Antibiotics.2021; 10(11): 1333. CrossRef - Association of phylogenetic distribution and presence of integrons with multidrug resistance in Escherichia coli clinical isolates from children with diarrhoea
Yesmi Patricia Ahumada-Santos, María Elena Báez-Flores, Sylvia Páz Díaz-Camacho, Magdalena de Jesús Uribe-Beltrán, Carlos Alberto Eslava-Campos, Jesús Ricardo Parra-Unda, Francisco Delgado-Vargas
Journal of Infection and Public Health.2020; 13(5): 767. CrossRef - Class 1 Integrons in Clinical Multi Drug Resistance E. coli, Sana’a Hospitals, Yemen
Mukhtar A. Al-Hammadi, Hassan A. Al-Shamahy, Abdulaziz Q. Ali, Mahfoudh A.M. Abdulghani, Hassan Pyar, Ibrahim AL-Suboal
Pakistan Journal of Biological Sciences.2020; 23(3): 231. CrossRef - Occurrence, Phenotypic and Molecular Characterization of Extended-Spectrum- and AmpC- β-Lactamase Producing Enterobacteriaceae Isolated From Selected Commercial Spinach Supply Chains in South Africa
Loandi Richter, Erika M. du Plessis, Stacey Duvenage, Lise Korsten
Frontiers in Microbiology.2020;[Epub] CrossRef - Staphylococcus aureus biofilms: Structures, antibiotic resistance, inhibition, and vaccines
Raziey Parastan, Mohammad Kargar, Kavous Solhjoo, Farshid Kafilzadeh
Gene Reports.2020; 20: 100739. CrossRef - Dissemination of antibiotic resistance genes (ARGs) via integrons in Escherichia coli: A risk to human health
Shaqiu Zhang, Muhammad Abbas, Mujeeb Ur Rehman, Yahui Huang, Rui Zhou, Siyue Gong, Hong Yang, Shuling Chen, Mingshu Wang, Anchun Cheng
Environmental Pollution.2020; 266: 115260. CrossRef Multi-Drug-Resistant Diarrheagenic Escherichia coli Pathotypes in Pediatric Patients with Gastroenteritis from Central Iran
Elnaz Abbasi, Mahdieh mondanizadeh, Alex van Belkum, Ehsanollah Ghaznavi-Rad
Infection and Drug Resistance.2020; Volume 13: 1387. CrossRef- The Relationship of Class I Integron Gene Cassettes and the Multidrug-Resistance in Extended -Spectrum β-Lactamase Producing Isolates of Escherichia coli
Alisha Akya, Roya Chegene Lorestani, Mosayeb Rostamian, Azam Elahi, Shokofe Baakhshii, Minoo Aliabadi, Keyghobad Ghadiri
Archives of Pediatric Infectious Diseases.2019;[Epub] CrossRef - Integrons in Enterobacteriaceae : diversity, distribution and epidemiology
Megha Kaushik, Sanjay Kumar, Rajeev Kumar Kapoor, Jugsharan Singh Virdi, Pooja Gulati
International Journal of Antimicrobial Agents.2018; 51(2): 167. CrossRef - Molecular analysis of Shiga toxin-producing Escherichia coli O157:H7 and non-O157 strains isolated from calves
Maryam Kohansal, Ali Ghanbari Asad
Onderstepoort Journal of Veterinary Research.2018;[Epub] CrossRef - Prevalence of Integrons and Insertion Sequences in ESBL-Producing E. coli Isolated from Different Sources in Navarra, Spain
Lara Pérez-Etayo, Melibea Berzosa, David González, Ana Vitas
International Journal of Environmental Research an.2018; 15(10): 2308. CrossRef - Frequency of antimicrobial resistance and integron gene cassettes in Escherichia coli isolated from giant pandas (Ailuropoda melanoleuca) in China
Wencheng Zou, Caiwu Li, Xin Yang, Yongxiang Wang, Guangyang Cheng, Jinxin Zeng, Xiuzhong Zhang, Yanpeng Chen, Run Cai, Qianru Huang, Lan Feng, Hongning Wang, Desheng Li, Guiquan Zhang, Yanxi Chen, Zhizhong Zhang, Heming Zhang
Microbial Pathogenesis.2018; 116: 173. CrossRef - Colistin-Resistant Klebsiella pneumoniae: Prevalence of Integrons and Synergistic Out Turn for Colistin-Meropenem
Prasanth Manohar, Thamaraiselvan Shanthini, Pandey Ekta, Mahesan J B, Kodiveri Muthukaliannan Gothandam, Bulent Bozdogan, Nachimuthu Ramesh
Archives of Clinical Infectious Diseases.2018;[Epub] CrossRef - Evaluation of integrons classes 1–3 in extended spectrum beta-lactamases and multi drug resistant Escherichia coli isolates in the North of Iran
Shahla Asgharzadeh Kangachar, Ali Mojtahedi
Gene Reports.2018; 12: 299. CrossRef - Association of Glycerol Kinase Gene with Class 3 Integrons: A Novel Cassette Array within Escherichia coli
Rajkumari Elizabeth, Debadatta Dhar Chanda, Atanu Chakravarty, Deepjyoti Paul, Shiela Chetri, Deepshikha Bhowmik, Jayalaxmi Wangkheimayum, Amitabha Bhattacharjee
Indian Journal of Medical Microbiology.2018; 36(1): 104. CrossRef - Emergence of class 1 to 3 integrons among members of Enterobacteriaceae in Egypt
Dina E. Rizk, Areej M. El-Mahdy
Microbial Pathogenesis.2017; 112: 50. CrossRef - Detection of Class I and II integrons for the assessment of antibiotic and multidrug resistance amongEscherichia coliisolates from agricultural irrigation waters in Bulacan, Philippines
Cielo Emar M. Paraoan, Windell L. Rivera, Pierangeli G. Vital
Journal of Environmental Science and Health, Part .2017; 52(5): 306. CrossRef - Distribution of Integrons and Phylogenetic Groups among Enteropathogenic Escherichia coli Isolates from Children <5 Years of Age in Delhi, India
Taru Singh, Shukla Das, V. G. Ramachandran, Sayim Wani, Dheeraj Shah, Khan A. Maroof, Aditi Sharma
Frontiers in Microbiology.2017;[Epub] CrossRef - The distribution of carbapenem- and colistin-resistance in Gram-negative bacteria from the Tamil Nadu region in India
Prasanth Manohar, Thamaraiselvan Shanthini, Ramankannan Ayyanar, Bulent Bozdogan, Aruni Wilson, Ashok J. Tamhankar, Ramesh Nachimuthu, Bruno S. Lopes
Journal of Medical Microbiology .2017; 66(7): 874. CrossRef - Multidrug-Resistant Escherichia coli Strains Isolated from Patients Are Associated with Class 1 and 2 Integrons
Hamid Lavakhamseh, Parviz Mohajeri, Samaneh Rouhi, Pegah Shakib, Rashid Ramazanzadeh, Afshin Rasani, Majid Mansouri
Chemotherapy.2016; 61(2): 72. CrossRef - Variability in gene cassette patterns of class 1 and 2 integrons associated with multi drug resistance patterns in Staphylococcus aureus clinical isolates in Tehran-Iran
Mahdi Mostafa, Seyed Davar Siadat, Fereshteh Shahcheraghi, Farzam Vaziri, Alireza Japoni-Nejad, Jalil Vand Yousefi, Bahareh Rajaei, Elnaz Harifi Mood, Nayyereh Ebrahim zadeh, Arfa Moshiri, Seyed Alireza Seyed Siamdoust, Mohamad Rahbar
BMC Microbiology.2015;[Epub] CrossRef
- Municipal wastewater treatment plant showing a potential reservoir for clinically relevant MDR bacterial strains co-occurrence of ESBL genes and integron-integrase genes
In Vitro Antibacterial Efficacy of 21 Indian Timber-Yielding Plants Against Multidrug-Resistant Bacteria Causing Urinary Tract Infection- Monali P. Mishra, Rabindra N. Padhy
- Osong Public Health Res Perspect. 2013;4(6):347-357. Published online December 31, 2013
- DOI: https://doi.org/10.1016/j.phrp.2013.10.007
- 3,057 View
- 17 Download
- 20 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
To screen methanolic leaf extracts of 21 timber-yielding plants for antibacterial activity against nine species of uropathogenic bacteria isolated from clinical samples of a hospital (Enterococcus faecalis, Staphylococcus aureus, Acinetobacter baumannii, Citrobacter freundii, Enterobacter aerogenes, Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, and Pseudomonas aeruginosa).
Methods
Bacterial strains were subjected to antibiotic sensitivity tests by the Kirby–Bauer's disc diffusion method. The antibacterial potentiality of leaf extracts was monitored by the agar-well diffusion method with multidrug-resistant (MDR) strains of nine uropathogens.
Results
Two Gram-positive isolates, E. faecalis and S. aureus, were resistant to 14 of the 18 antibiotics used. Gram-negative isolates A. baumannii, C. freundii, E. aerogenes, E. coli, K. pneumoniae, P. mirabilis, and P. aeruginosa were resistant to 10, 12, 9, 11, 11, 10, and 11 antibiotics, respectively, of the 14 antibiotics used. Methanolic leaf extracts of Anogeissus acuminata had the maximum zone of inhibition size—29 mm against S. aureus and 28 mm against E. faecalis and P. aeruginosa. Cassia tora had 29 mm as the zone of inhibition size for E. faecalis, E. aerogenes, and P. aeruginosa. Based on the minimum inhibitory concentration and minimum bactericidal concentration values, the most effective 10 plants against uropathogens could be arranged in decreasing order as follows: C. tora > A. acuminata > Schleichera oleosa > Pterocarpus santalinus > Eugenia jambolana > Bridelia retusa > Mimusops elengi > Stereospermum kunthianum > Tectona grandis > Anthocephalus cadamba. The following eight plants had moderate control capacity: Artocarpus heterophyllus, Azadirachta indica, Dalbergia latifolia, Eucalyptus citriodora, Gmelina arborea, Pongamia pinnata, Pterocarpus marsupium, and Shorea robusta. E. coli, followed by A. baumannii, C. freundii, E. aerogenes, P. mirabilis, and P. aeruginosa were controlled by higher amounts/levels of leaf extracts. Phytochemicals of all plants were qualitatively estimated.
Conclusions
A majority of timber-yielding plants studied had in vitro control capacity against MDR uropathogenic bacteria. -
Citations
Citations to this article as recorded by- Chemical constituents, pharmacological activities, and uses of common ayurvedic medicinal plants: a future source of new drugs
Parul Kaushik, Priyanka Ahlawat, Kuldeep Singh, Raman Singh
Advances in Traditional Medicine.2023; 23(3): 673. CrossRef - Antibacterial activity of Libyan Juniperus phoenicea L. leaves extracts against common nosocomial pathogens
Aml O. Alhadad, Galal S. Salem, Suliman M. Hussein, Sarah M. Elshareef
Journal of Experimental Biology and Agricultural S.2023; 11(2): 371. CrossRef - The Ethnopharmacology, Phytochemistry and Bioactivities of the Corymbia Genus (Myrtaceae)
Matthew J. Perry, Phurpa Wangchuk
Plants.2023; 12(21): 3686. CrossRef - Gas Chromatography-Mass Spectroscopic, high performance liquid chromatographic and In-silico characterization of antimicrobial and antioxidant constituents of Rhus longipes(Engl)
Adedoyin Adetutu Olasunkanmi, Olumide Samuel Fadahunsi, Peter Ifeoluwa Adegbola
Arabian Journal of Chemistry.2022; 15(2): 103601. CrossRef - Evaluation of the sensory attributes of pepper soup beef hides and determination of the preservative potential of the spices used for its preparation
T.C.L. Maguipa, P.D. Mbougueng, H.M. Womeni
Journal of Agriculture and Food Research.2022; 8: 100293. CrossRef - Flourensia retinophylla: An outstanding plant from northern Mexico with antibacterial activity
D. Jasso de Rodríguez, M.C. Victorino-Jasso, N.E. Rocha-Guzmán, M.R. Moreno-Jiménez, L. Díaz-Jiménez, R. Rodríguez-García, J.Á. Villarreal-Quintanilla, F.M. Peña-Ramos, D.A. Carrillo-Lomelí, Z.A. Genisheva, M.L. Flores-López
Industrial Crops and Products.2022; 185: 115120. CrossRef - Antibacterial Screening, Biochemometric and Bioautographic Evaluation of the Non-Volatile Bioactive Components of Three Indigenous South African Salvia Species
Margaux Lim Ah Tock, Sandra Combrinck, Guy Kamatou, Weiyang Chen, Sandy Van Vuuren, Alvaro Viljoen
Antibiotics.2022; 11(7): 901. CrossRef - Computational in Silico Modelling of Phytochemicals as a Potential Cure
Rachita Kurmi, Kavya N R, Jennath Sherin A, Silpa T S
International Journal of Scientific Research in Sc.2021; : 42. CrossRef - Hepatoprotective effects of Cassiae Semen on mice with non-alcoholic fatty liver disease based on gut microbiota
Hanyan Luo, Hongwei Wu, Lixia Wang, Shuiming Xiao, Yaqi Lu, Cong Liu, Xiankuo Yu, Xiao Zhang, Zhuju Wang, Liying Tang
Communications Biology.2021;[Epub] CrossRef - Phytochemical screening, anthocyanins and antimicrobial activities in some berries fruits
Amina A. Aly, Hoda G. M. Ali, Noha E. R. Eliwa
Journal of Food Measurement and Characterization.2019; 13(2): 911. CrossRef - Antimicrobial activity of select edible plants from Odisha, India against food-borne pathogens
Sujogya Kumar Panda, Yugal Kishore Mohanta, Laxmipriya Padhi, Walter Luyten
LWT.2019; 113: 108246. CrossRef - Extracts of Tectona grandis and Vernonia amygdalina have anti-Toxoplasma and pro-inflammatory properties in vitro
Mlatovi Dégbé, Françoise Debierre-Grockiego, Amivi Tété-Bénissan, Héloïse Débare, Kodjo Aklikokou, Isabelle Dimier-Poisson, Messanvi Gbeassor
Parasite.2018; 25: 11. CrossRef - Psacalium paucicapitatum has in vitro antibacterial activity
D. Jasso de Rodríguez, L.C. García-Hernández, N.E. Rocha-Guzmán, M.R. Moreno-Jiménez, R. Rodríguez-García, M.L.V. Díaz-Jiménez, A. Sáenz-Galindo, J.A. Villarreal-Quintanilla, F.M. Peña-Ramos, M.L. Flores-López, D.A. Carrillo-Lomelí
Industrial Crops and Products.2017; 107: 489. CrossRef - Qualitative Phytochemical Analysis, Antimicrobial Activity and Cytotoxic Effect of Moringa concanensis Nimmo Leaves
Ramaswamy Malathi, Solaimuthu Chandrasek
Research Journal of Medicinal Plants.2017; 11(3): 93. CrossRef - In vitro antibacterial activity of crude extracts of 9 selected medicinal plants against UTI causing MDR bacteria
Monali P. Mishra, Sibanarayan Rath, Shasank S. Swain, Goutam Ghosh, Debajyoti Das, Rabindra N. Padhy
Journal of King Saud University - Science.2017; 29(1): 84. CrossRef - Phytochemical investigation and antimicrobial assessment of Bellis sylvestris leaves
Monica Scognamiglio, Elisabetta Buommino, Lorena Coretti, Vittoria Graziani, Rosita Russo, Pina Caputo, Giovanna Donnarumma, Brigida D⿿Abrosca, Antonio Fiorentino
Phytochemistry Letters.2016; 17: 6. CrossRef - The genus Anogeissus: A review on ethnopharmacology, phytochemistry and pharmacology
Deeksha Singh, Uttam Singh Baghel, Anshoo Gautam, Dheeraj Singh Baghel, Divya Yadav, Jai Malik, Rakesh Yadav
Journal of Ethnopharmacology.2016; 194: 30. CrossRef - Antibacterial activity of five Peruvian medicinal plants against Pseudomonas aeruginosa
Gabriela Ulloa-Urizar, Miguel Angel Aguilar-Luis, María del Carmen De Lama-Odría, José Camarena-Lizarzaburu, Juana del Valle Mendoza
Asian Pacific Journal of Tropical Biomedicine.2015; 5(11): 928. CrossRef - In vitro antibacterial efficacy of plants used by an Indian aboriginal tribe against pathogenic bacteria isolated from clinical samples
Shasank S. Swain, Rabindra N. Padhy
Journal of Taibah University Medical Sciences.2015; 10(4): 379. CrossRef - Monitoring in vitro antibacterial efficacy of 26 Indian spices against multidrug resistant urinary tract infecting bacteria
Sibanarayan Rath, Rabindra N. Padhy
Integrative Medicine Research.2014; 3(3): 133. CrossRef
- Chemical constituents, pharmacological activities, and uses of common ayurvedic medicinal plants: a future source of new drugs
- Comparison of Antimicrobial Resistance in
Escherichia coli Strains Isolated From Healthy Poultry and Swine Farm Workers Using Antibiotics in Korea - Seung-Hak Cho, Yeong-Sik Lim, Yeon-Ho Kang
- Osong Public Health Res Perspect. 2012;3(3):151-155. Published online June 30, 2012
- DOI: https://doi.org/10.1016/j.phrp.2012.07.002
- 2,788 View
- 21 Download
- 22 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
The aim of this study is to compare the antibiotic resistance of Escherichia coli isolates from faecal samples of workers who often use antibiotics.
Methods
A total of 163E coli strains isolated from faecal samples of livestock workers (poultry and swine farm workers) and restaurant workers in the same regions as a control group were analyzed by agar disc diffusion to determine their susceptibility patterns to 16 antimicrobial agents.
Results
Most of the tested isolates showed high antimicrobial resistance to ampicillin and tetracycline. The isolates showed higher resistance to cephalothin than other antibiotics among the cephems. Among the aminoglycosides, the resistance to gentamicin and tobramycin occurred at higher frequencies compared with resistance to amikacin and netilmicin. Our data indicated that faecal E coli isolates of livestock workers showed higher antibiotic resistances than nonlivestock workers (restaurant workers), especially cephalothin, gentamicin, and tobramycin (p < 0.05). Moreover, the rates of the livestock workers in the association of multidrug resistance were also higher than the rates of the restaurant workers.
Conclusion
This study implies that usage of antibiotics may contribute to the prevalence of antibiotic resistance in commensal E coli strains of humans. -
Citations
Citations to this article as recorded by- Comparison of Antimicrobial Resistance in Escherichia coli Strains Isolated From Swine, Poultry, and Farm Workers in the Respective Livestock Farming Units in Greece
Magdalini K Christodoulou
Cureus.2023;[Epub] CrossRef - Risk Factors Associated with the Carriage of Pathogenic Escherichia coli in Healthy Commercial Meat Chickens in Queensland, Australia †
Leena Awawdeh, Rachel Forrest, Conny Turni, Rowland Cobbold, Joerg Henning, Justine Gibson
Poultry.2022; 1(2): 94. CrossRef - Characteristics and nutrient function of intestinal bacterial communities in black soldier fly (Hermetia illucens L.) larvae in livestock manure conversion
Yue Ao, Chongrui Yang, Shengchen Wang, Qingyi Hu, Li Yi, Jibin Zhang, Ziniu Yu, Minmin Cai, Chan Yu
Microbial Biotechnology.2021; 14(3): 886. CrossRef - Insects, Rodents, and Pets as Reservoirs, Vectors, and Sentinels of Antimicrobial Resistance
Willis Gwenzi, Nhamo Chaukura, Norah Muisa-Zikali, Charles Teta, Tendai Musvuugwa, Piotr Rzymski, Akebe Luther King Abia
Antibiotics.2021; 10(1): 68. CrossRef - Phenotypic and genotypic antimicrobial resistance patterns of Escherichia coli and Klebsiella isolated from dairy farm milk, farm slurry and water in Punjab, India
Prateek Jindal, Jasbir Bedi, Randhir Singh, Rabinder Aulakh, Jatinder Gill
Environmental Science and Pollution Research.2021; 28(22): 28556. CrossRef - Analysis of drug sensitivity of Escherichia Coli O157H7
Minzi Xu, Zhenyu Liu, Yanbo Song, Runan Zhao, Zheng Yang, Huijin Zhao, Xiaobing Sun, Yaning Gu, Huifei Yang
Biomedical Microdevices.2021;[Epub] CrossRef - One Health of Peripheries: Biopolitics, Social Determination, and Field of Praxis
Oswaldo Santos Baquero
Frontiers in Public Health.2021;[Epub] CrossRef - Antimicrobial resistance in fecal Escherichia coli isolated from poultry chicks in northern Iran
Zohreh Pourhossein, Leila Asadpour, Hadi Habibollahi, Seyedeh Tooba Shafighi
Gene Reports.2020; 21: 100926. CrossRef - Prevalence of Multidrug Resistant Escherichia Coli In Suspected Cases of Urinary Tract Infection Among Patients Attending Ahmadu Bello University Medical Center, Zaria
Shitu, S., Gambo, B. A., Musa, M.O., Abubakar, A.A., Attahiru, M.
UMYU Journal of Microbiology Research (UJMR).2020; 5(2): 123. CrossRef - Prevalence and risk factors for multi-drug resistant Escherichia coli among poultry workers in the Federal Capital Territory, Abuja, Nigeria
Mabel Kamweli Aworh, Jacob Kwaga, Emmanuel Okolocha, Nwando Mba, Siddhartha Thakur, Grzegorz Woźniakowski
PLOS ONE.2019; 14(11): e0225379. CrossRef - High genomic diversity of multi-drug resistant wastewater Escherichia coli
Norhan Mahfouz, Serena Caucci, Eric Achatz, Torsten Semmler, Sebastian Guenther, Thomas U. Berendonk, Michael Schroeder
Scientific Reports.2018;[Epub] CrossRef - Characterization of Escherichia coli Isolated from Day-old Chicken Fluff in Taiwanese Hatcheries
Shengnan Zhao, Chia-Lan Wang, Shao-Kuang Chang, Yi-Lun Tsai, Chung-Hsi Chou
Avian Diseases.2018; 63(1): 9. CrossRef - Changes in antimicrobial resistance patterns and dominance of extended spectrum β-lactamase genes among faecal Escherichia coli isolates from broilers and workers during two rearing periods
Fatemeh Doregiraee, Masoud Alebouyeh, Bahar Nayeri Fasaei, Saeed Charkhkar, Elahe Tajeddin, Mohammad Reza Zali
Italian Journal of Animal Science.2018; 17(3): 815. CrossRef - Oxytetracycline reduces the diversity of tetracycline-resistance genes in the Galleria mellonella gut microbiome
Katarzyna Ignasiak, Anthony Maxwell
BMC Microbiology.2018;[Epub] CrossRef - Antimicrobial Resistance and the Presence of Virulence Genes in Escherichia coli Strains Isolated from Ruditapes philippinarum in Gomso Bay, Korea
Tae-Ok Kim, In-Seon Eom, Kwang-Ho Park, Kwon-Sam Park
Korean Journal of Fisheries and Aquatic Sciences.2016; 49(6): 800. CrossRef - High prevalence of cross-resistance to fluoroquinolone and cotrimoxazole in tetracycline-resistant Escherichia coli human clinical isolates
Eric Batard, Mathilde Lefebvre, Guillaume Ghislain Aubin, Nathalie Caroff, Stéphane Corvec
Journal of Chemotherapy.2016; 28(6): 510. CrossRef - Soil‐borne reservoirs of antibiotic‐resistant bacteria are established following therapeutic treatment of dairy calves
Jinxin Liu, Zhe Zhao, Lisa Orfe, Murugan Subbiah, Douglas R. Call
Environmental Microbiology.2016; 18(2): 557. CrossRef - Antibiotic Resistance in Airborne Bacteria Near Conventional and Organic Beef Cattle Farms in California, USA
Helen M. Sancheza, Cristina Echeverria, Vanessa Thulsiraj, Amy Zimmer-Faust, Ariel Flores, Madeleine Laitz, Gregory Healy, Shaily Mahendra, Suzanne E. Paulson, Yifang Zhu, Jennifer A. Jay
Water, Air, & Soil Pollution.2016;[Epub] CrossRef - Transport of Antibiotic Resistance Plasmids in Porous Media
Chaoqi Chen, Jing Li, Stephanie L. DeVries, Pengfei Zhang, Xiqing Li
Vadose Zone Journal.2015; 14(3): 1. CrossRef - Possibility of CTX-M-14 Gene Transfer from Shigella sonnei to a Commensal Escherichia coli Strain of the Gastroenteritis Microbiome
Seung-Hak Cho, Soon Young Han, Yeon-Ho Kang
Osong Public Health and Research Perspectives.2014; 5(3): 156. CrossRef - A survey of the frequency of aminoglycoside antibiotic-resistant genotypes and phenotypes inEscherichia coliin broilers with septicaemia in Hebei, China
F.Y. Zhang, S.Y. Huo, Y.R. Li, R. Xie, X.J. Wu, L.G. Chen, Y.H. Gao
British Poultry Science.2014; 55(3): 305. CrossRef - Prevalence of Antimicrobial Resistance in Escherichia coli Strains Isolated from Fishery Workers
Hyun-Ho Shin, Seung-Hak Cho
Osong Public Health and Research Perspectives.2013; 4(2): 72. CrossRef
- Comparison of Antimicrobial Resistance in Escherichia coli Strains Isolated From Swine, Poultry, and Farm Workers in the Respective Livestock Farming Units in Greece
- The Emergence of Oseltamivir-Resistant Seasonal Influenza A (H1N1) Virus in Korea During the 2008-2009 Season
- Woo-Young Choi, Inseok Yang, Sujin Kim, Namjoo Lee, Meehwa Kwon, Joo-Yeon Lee, Chun Kang
- Osong Public Health Res Perspect. 2011;2(3):178-185. Published online December 31, 2011
- DOI: https://doi.org/10.1016/j.phrp.2011.11.042
- 2,724 View
- 14 Download
- 10 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
To monitor antiviral drug resistance among seasonal influenza viruses isolated in Korea during the 2008-2009 influenza season, we examined influenza isolates collected through Korea Influenza Surveillance Scheme for antiviral drug susceptibility.
Methods
For genetic analysis of antiviral drug resistance, the matrix (M2) and neuraminidase (NA) genes of each isolate were amplified by reverse transcription-polymerase chain reaction and followed by nucleotide sequencing. For phylogenetic analyses, the sequences of hemagglutinin (HA) and NA genes of each isolate were aligned using multiple alignment program. For phenotypic analysis of antiviral drug resistance, drug susceptibilities against M2 inhibitor (amantadine) and NA inhibitors (oseltavimir and zanamivir) were determined by virus yield reduction assay and fluorometric NA inhibition assay, respectively.
Results
In Korea, the resistant influenza viruses against oseltamivir were first detected in sealsonal influenza A(H1N1) viruses on Week 48 of 2008. Since then, the number of oseltamivir-resistant A(H1N1) viruses was continuously increased and had reached the highest peak on Week 52 of 2008. 533 (99.8%) of 534 A(H1N1) viruses were resistant to oseltamivir and all of them harbored the H275Y mutation in the NA gene during the 2008-2009 season. The oseltamivir resistance identified by sequencing was confirmed by NA inhibition assay. Genetic analysis based on HA gene of the resistant A(H1N1) viruses revealed that the viruses were identified as A/Brisbane/10/2007-like strain which was vaccine strain for the 2008-2009 season.
Conclusions
The oseltamivir-resistant A(H1N1) viruses were first emerged in Europe in November 2007 and then circulated globally. One year later, the oseltamivir-resistant A(H1N1) viruses were first detected in Korea in November 2008 and continued circulating until the Week 7 of 2009 during the 2008-2009 season. Considering the pandemic preparedness, it should be continued to monitor the emergence and the characterization of antiviral drug resistant influenza viruses. -
Citations
Citations to this article as recorded by- Pharmacokinetics and safety of a novel influenza treatment (baloxavir marboxil) in Korean subjects compared with Japanese subjects
Yun Kim, Sangwon Lee, Yohan Kim, In‐Jin Jang, SeungHwan Lee
Clinical and Translational Science.2022; 15(2): 422. CrossRef - 2018–2019 antiviral drug sensitivity of the influenza virus strains isolated from various regions of Kazakhstan
T. I. Glebova, N. G. Klivleyeva, G. V. Lukmanova, N. T. Saktaganov, A. M. Baimukhametova
Russian Journal of Infection and Immunity.2021; 11(6): 1159. CrossRef - Assessment of Intensive Vaccination and Antiviral Treatment in 2009 Influenza Pandemic in Korea
Chaeshin Chu, Sunmi Lee
Osong Public Health and Research Perspectives.2015; 6(1): 47. CrossRef - Doing Mathematics with Aftermath of Pandemic Influenza 2009
Hae-Wol Cho, Chaeshin Chu
Osong Public Health and Research Perspectives.2015; 6(1): 1. CrossRef - Antiviral treatment of influenza in South Korea
Young June Choe, Hyunju Lee, Hoan Jong Lee, Eun Hwa Choi
Expert Review of Anti-infective Therapy.2015; 13(6): 741. CrossRef - Synthesis and anti-influenza virus activity of 4-oxo- or thioxo-4,5-dihydrofuro[3,4-c]pyridin-3(1H)-ones
Ye Jin Jang, Raghavendra Achary, Hye Won Lee, Hyo Jin Lee, Chong-Kyo Lee, Soo Bong Han, Young-Sik Jung, Nam Sook Kang, Pilho Kim, Meehyein Kim
Antiviral Research.2014; 107: 66. CrossRef - Was the Mass Vaccination Effective During the Influenza Pandemic 2009–2010 in Korea?
Hae-Wol Cho, Chaeshin Chu
Osong Public Health and Research Perspectives.2013; 4(4): 177. CrossRef - How to Manage a Public Health Crisis and Bioterrorism in Korea
Hae-Wol Cho, Chaeshin Chu
Osong Public Health and Research Perspectives.2013; 4(5): 223. CrossRef - Generation and Characterization of Recombinant Influenza A(H1N1) Viruses Resistant to Neuraminidase Inhibitors
WooYoung Choi, Jin-Young Shin, Hwan-Eui Jeong, Mi-Jin Jeong, Su-Jin Kim, Joo-Yeon Lee, Chun Kang
Osong Public Health and Research Perspectives.2013; 4(6): 323. CrossRef - Occurrence and characterization of oseltamivir-resistant influenza virus in children between 2007-2008 and 2008-2009 seasons
Seoung Geun Kim, Yoon Ha Hwang, Yung Hae Shin, Sung Won Kim, Woo Sik Jung, Sung Mi Kim, Jae Min Oh, Na Young Lee, Mun Ju Kim, Kyung Soon Cho, Yeon Gyeong Park, Sang Kee Min, Chang Kyu Lee, Jun Sub Kim, Chun Kang, Joo Yeon Lee, Man Kyu Huh, Chang Hoon Kim
Korean Journal of Pediatrics.2013; 56(4): 165. CrossRef
- Pharmacokinetics and safety of a novel influenza treatment (baloxavir marboxil) in Korean subjects compared with Japanese subjects
- Prevalence of Antibiotic Resistance in
Escherichia coli Fecal Isolates From Healthy Persons and Patients With Diarrhea - Seung-Hak Cho, Yeong-Sik Lim, Mi-Sun Park, Seong-Han Kim, Yeon-Ho Kang
- Osong Public Health Res Perspect. 2011;2(1):41-45. Published online June 30, 2011
- DOI: https://doi.org/10.1016/j.phrp.2011.05.003
- 2,842 View
- 20 Download
- 11 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
This study aimed to investigate the prevalence of antibiotic resistance in fecal Escherichia coli isolates from healthy persons and patients with diarrhea.
Methods
E. coli isolates (n = 428) were obtained from fecal samples of apparently healthy volunteers and hospitalized patients with diarrhea. Susceptibility patterns of isolates to 16 antimicrobial agents were determined by agar disc diffusion.
Results
Most E. coli isolates exhibited less than 10% resistance against imipenem, cefotetan, aztreonam, cefepime, cefoxitin, amikacin and netilamicin, although greater than 65% were resistant to ampicillin and tetracycline. No significant difference in resistance rates for all tested antibiotics was found between isolates from the healthy-and diarrheal-patient groups, including for multi-drug resistance (p = 0.22). The highest number of resistant antibiotics was 12 antibiotics. No significant differences in antibiotic resistance were found among the sex and age strata for isolates from healthy individuals. However, antibiotic resistance rates to cefoxitin, cefotaxime, amikacin, and netilamicin were significantly higher in the isolates of men than those of women (p < 0.05) in isolates from patients with diarrhea. Furthermore, isolates from patients with diarrhea older than 40-years of age showed higher resistance to cefepime and aztreonam (p < 0.05).
Conclusion
High resistance to the antibiotics most frequently prescribed for diarrhea was found in isolates from patients with diarrhea and apparently healthy individuals without any significant difference. -
Citations
Citations to this article as recorded by- Characterization and antimicrobial susceptibility of Escherichia coli isolated from healthy farm animals in Tunisia
Salma Bessalah, John Morris Fairbrother, Imed Salhi, Ghyslaine Vanier, Touhami Khorchani, Mabrouk-Mouldi Seddik, Mohamed Hammadi
Animal Biotechnology.2021; 32(6): 748. CrossRef - Research note: Occurrence ofmcr-encoded colistin resistance inEscherichia colifrom pigs and pig farm workers in Vietnam
Son Thi Thanh Dang, Duong Thi Quy Truong, John Elmerdahl Olsen, Nhat Thi Tran, Giang Thi Huong Truong, Hue Thi Kim Vu, Anders Dalsgaard
FEMS Microbes.2021;[Epub] CrossRef - Multidrug-resistant bacteria as intestinal colonizers and evolution of intestinal colonization in healthy university students in Portugal
Raquel Mota, Marisa Pinto, Josman Palmeira, Daniela Gonçalves, Helena Ferreira
Access Microbiology .2021;[Epub] CrossRef - Influence of Environmental and Anthropogenic Factors on Microbial Ecology and Sanitary Threat in the Final Stretch of the Brda River
Łukasz Kubera, Marta Małecka-Adamowicz, Emilia Jankowiak, Ewa Dembowska, Piotr Perliński, Karolina Hejze
Water.2019; 11(5): 922. CrossRef - Comparison of Commensal Escherichia coli Isolates from Adults and Young Children in Lubuskie Province, Poland: Virulence Potential, Phylogeny and Antimicrobial Resistance
Ewa Bok, Justyna Mazurek, Andrzej Myc, Michał Stosik, Magdalena Wojciech, Katarzyna Baldy-Chudzik
International Journal of Environmental Research an.2018; 15(4): 617. CrossRef - PHENOTYPIC DETECTION OF AMPC β-LACTAMASE ENZYME IN GRAM-NEGATIVE BACILLI
Khanda Anoar, Sherko Omer, Bayan Majid, Hero Rahim, Shno Muhammed
JOURNAL OF SULAIMANI MEDICAL COLLEGE.2018; 8(2): 57. CrossRef - Antimicrobial resistance profiles and molecular characterization of Escherichia coli strains isolated from healthy adults in Ho Chi Minh City, Vietnam
Phuong Hoai HOANG, Sharda Prasad AWASTHI, Phuc DO NGUYEN, Ngan Ly Hoang NGUYEN, Dao Thi Anh NGUYEN, Ninh Hoang LE, Chinh VAN DANG, Atsushi HINENOYA, Shinji YAMASAKI
Journal of Veterinary Medical Science.2017; 79(3): 479. CrossRef - Characterization of enteropathogenicEscherichia coliof clinical origin from the pediatric population in Pakistan
Mahwish Younas, Fariha Siddiqui, Zobia Noreen, Syeda Sadia Bokhari, Oscar G. Gomez-Duarte, Brendan W. Wren, Habib Bokhari
Transactions of The Royal Society of Tropical Medi.2016; 110(7): 414. CrossRef - Possibility of CTX-M-14 Gene Transfer from Shigella sonnei to a Commensal Escherichia coli Strain of the Gastroenteritis Microbiome
Seung-Hak Cho, Soon Young Han, Yeon-Ho Kang
Osong Public Health and Research Perspectives.2014; 5(3): 156. CrossRef - Prevalence of Antimicrobial Resistance in Escherichia coli Strains Isolated from Fishery Workers
Hyun-Ho Shin, Seung-Hak Cho
Osong Public Health and Research Perspectives.2013; 4(2): 72. CrossRef - The Road Less Traveled
Chaeshin Chu
Osong Public Health and Research Perspectives.2011; 2(1): 1. CrossRef
- Characterization and antimicrobial susceptibility of Escherichia coli isolated from healthy farm animals in Tunisia



 First
First Prev
Prev


