Search
- Page Path
- HOME > Search
- Drug resistance and the genotypic characteristics of rpoB and katG in rifampicin- and/or isoniazid-resistant Mycobacterium tuberculosis isolates in central Vietnam
- Thi Binh Nguyen Nguyen, Thi Kieu Diem Nguyen, Van Hue Trương, Thi Tuyet Ngoc Tran, van Bao Thang Phan, Thi Tuyen Nguyen, Hoang Bach Nguyen, Viet Quynh Tram Ngo, Van Tuan Mai, Paola Molicotti
- Osong Public Health Res Perspect. 2023;14(5):347-355. Published online October 18, 2023
- DOI: https://doi.org/10.24171/j.phrp.2023.0124
- 1,126 View
- 76 Download
-
 Graphical Abstract
Graphical Abstract
 Abstract
Abstract
 PDF
PDF 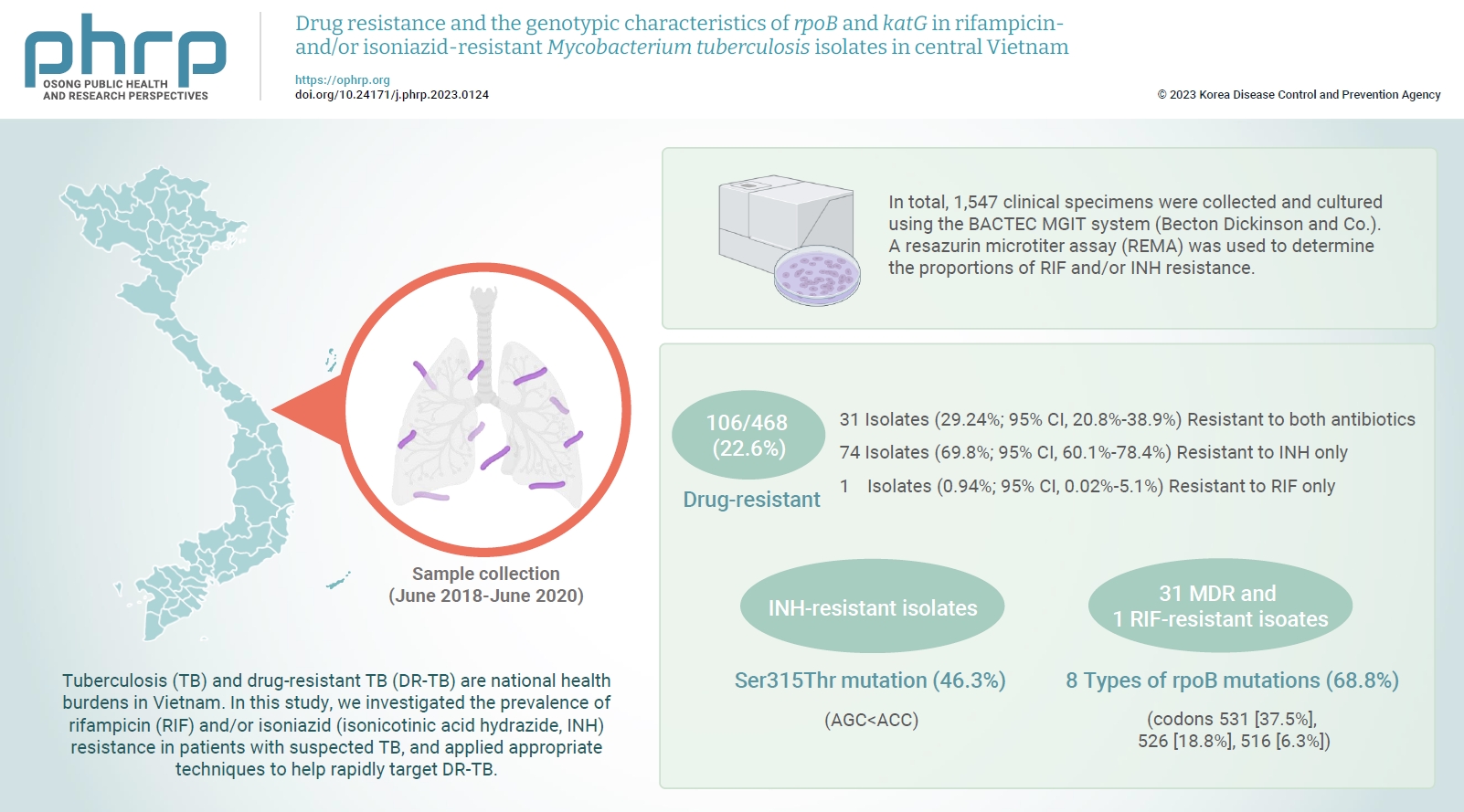
- Objectives
Tuberculosis (TB) and drug-resistant TB (DR-TB) are national health burdens in Vietnam. In this study, we investigated the prevalence of rifampicin (RIF) and/or isoniazid (isonicotinic acid hydrazide, INH) resistance in patients with suspected TB, and applied appropriate techniques to help rapidly target DR-TB. Methods: In total, 1,547 clinical specimens were collected and cultured using the BACTEC MGIT system (Becton Dickinson and Co.). A resazurin microtiter assay (REMA) was used to determine the proportions of RIF and/or INH resistance. A real-time polymerase chain reaction panel with TaqMan probes was employed to identify the mutations of rpoB and katG associated with DR-TB in clinical isolates. Genotyping of the identified mutations was also performed. Results: A total of 468 Mycobacterium tuberculosis isolates were identified using the REMA. Of these isolates, 106 (22.6%) were found to be resistant to 1 or both antibiotics. Of the resistant isolates, 74 isolates (69.8%) were resistant to isoniazid (INH) only, while 1 isolate (0.94%) was resistant to RIF only. Notably, 31 isolates (29.24%) were resistant to both antibiotics. Of the 41 phenotypically INH-resistant isolates, 19 (46.3%) had the Ser315Thr mutation. There were 8 different rpoB mutations in 22 (68.8%) of the RIF-resistant isolates. The most frequently detected mutations were at codons 531 (37.5%), 526 (18.8%), and 516 (6.3%). Conclusion: To help prevent new cases of DR-TB in Vietnam, it is crucial to gain a comprehensive understanding of the genotypic DR-TB isolates.
- Clinical epidemiological applicability of real-time polymerase chain reaction for COVID-19
- Geehyuk Kim, Jun-Kyu Kang, Jungho Kim, Jiyoung Lee, Jin Gwack
- Osong Public Health Res Perspect. 2022;13(4):252-262. Published online July 27, 2022
- DOI: https://doi.org/10.24171/j.phrp.2022.0135
- 3,272 View
- 160 Download
-
 Graphical Abstract
Graphical Abstract
 Abstract
Abstract
 PDF
PDF 
- Objectives
Real-time polymerase chain reaction is currently used as a confirmatory test for coronavirus disease 2019 (COVID-19). The test results are interpreted as positive, negative, or inconclusive, and are used only for a qualitative classification of patients. However, the test results can be quantitated using threshold count (Ct) values to determine the amount of virus present in the sample. Therefore, this study investigated the diagnostic usefulness of Ct results through various quantitative analyzes, along with an analysis of clinical and epidemiological characteristics.
Methods
Clinical and epidemiological data from 4,642 COVID-19 patients in April 2021 were analyzed, including the Ct values of the RNA-dependent RNA polymerase (RdRp), envelope (E), and nucleocapsid (N) genes. Clinical and epidemiological data (sex, age, underlying diseases, and early symptoms) were collected through a structured questionnaire. A correlation analysis was used to examine the relationships between variables.
Results
All 3 genes showed statistically significant relationships with symptoms and severity levels. The Ct values of the RdRp gene decreased as the severity of the patients increased. Moreover, statistical significance was observed for the presence of underlying diseases and dyspnea.
Conclusion
Ct values were found to be related to patients’ clinical and epidemiological characteristics. In particular, since these factors are closely related to symptoms and severity, Ct values can be used as primary data for predicting patients’ disease prognosis despite the limitations of this method. Conducting follow-up studies to validate this approach might enable using the data from this study to establish policies for preventing COVID-19 infection and spread.
- Voluntary testing for COVID-19: perceptions and utilization among the inhabitants of Saudi Arabia
- Ehab A. Abo-Ali, Ahmed Mousa, Rania Hussien, Shahad Mousa, Shayma Al-Rubaki, Mennatulla Omar, Badr Al-Haffashi, Abdullah Almilaibary
- Osong Public Health Res Perspect. 2022;13(3):212-220. Published online June 10, 2022
- DOI: https://doi.org/10.24171/j.phrp.2022.0062
- 3,143 View
- 86 Download
-
 Abstract
Abstract
 PDF
PDF - Objectives
Voluntary testing (VT) plays a crucial role in the prevention and control of infectious diseases. The present study investigated the perceptions and utilization of VT services for coronavirus disease 2019 (COVID-19) among the inhabitants of Saudi Arabia. Methods: In total, 3,510 adult participants from all provinces of Saudi Arabia were recruited via a national online survey. Results: Of the 3,510 participants, 88.9% were aware of the testing services available to them and of those, more than half (59.5%) had used the VT services and 96.1% were satisfied with the services. Contact with a positive COVID-19 case was the top reason for accessing VT, while a lack of awareness about the availability of VT services was the top perceived limiting factor. A history of chronic health conditions, anxiety and/or depression, and previous symptoms suggestive of COVID-19 were found to be predictors of the utilization of VT services (odds ratio [OR] 1.55, 95% confidence interval [CI] 1.22−1.96; OR 1.48, 95% CI 1.16−1.88; and OR 3.31, 95% CI 2.77−3.95), respectively. Conclusion: The awareness of voluntary COVID-19 testing services was satisfactory among the Saudi Arabian population, but can be improved. Sociodemographic and health history predictors of the utilization of VT services were identified.
- Specification of Bacteriophage Isolated Against Clinical Methicillin-Resistant
Staphylococcus Aureus - Ahmad Nasser, Reza Azizian, Mohsen Tabasi, Jamil Kheirvari Khezerloo, Fatemah Sadeghpour Heravi, Morovat Taheri Kalani, Norkhoda Sadeghifard, Razieh Amini, Iraj Pakzad, Amin Radmanesh, Farid Azizi Jalilian
- Osong Public Health Res Perspect. 2019;10(1):20-24. Published online February 28, 2019
- DOI: https://doi.org/10.24171/j.phrp.2019.10.1.05
- 8,680 View
- 71 Download
- 15 Crossref
-
 Abstract
Abstract
 PDF
PDF Objectives The emergence of resistant bacteria is being increasingly reported around the world, potentially threatening millions of lives. Amongst resistant bacteria, methicillin-resistant
Staphylococcus aureus (MRSA) is the most challenging to treat. This is due to emergent MRSA strains and less effective traditional antibiotic therapies to Staphylococcal infections. The use of bacteriophages (phages) against MRSA is a new, potential alternate therapy. In this study, morphology, genetic and protein structure of lytic phages against MRSA have been analysed.Methods Isolation of livestock and sewage bacteriophages were performed using 0.4 μm membrane filters. Plaque assays were used to determine phage quantification by double layer agar method. Pure plaques were then amplified for further characterization. Sulfate-polyacrylamide gel electrophoresis and random amplification of polymorphic DNA were run for protein evaluation, and genotyping respectively. Transmission electron microscope was also used to detect the structure and taxonomic classification of phage visually.
Results Head and tail morphology of bacteriophages against MRSA were identified by transmission electron microscopy and assigned to the
Siphoviridae family and theCaudovirales order.Conclusion Bacteriophages are the most abundant microorganism on Earth and coexist with the bacterial population. They can destroy bacterial cells successfully and effectively. They cannot enter mammalian cells which saves the eukaryotic cells from lytic phage activity. In conclusion, phage therapy may have many potential applications in microbiology and human medicine with no side effect on eukaryotic cells.
-
Citations
Citations to this article as recorded by- Characterization of a Vibrio parahaemolyticus-targeting lytic bacteriophage SSJ01 and its application in artificial seawater
Jungu Kang, Yoonjee Chang
Food Science and Biotechnology.2024; 33(6): 1505. CrossRef - Isolation and encapsulation of bacteriophage with chitosan nanoparticles for biocontrol of multidrug-resistant methicillin-resistant Staphylococcus aureus isolated from broiler poultry farms
Mona M. Elsayed, Rasha M. Elkenany, Ayman Y. EL-Khateeb, Nehal M. Nabil, Maram M. Tawakol, Heba M. Hassan
Scientific Reports.2024;[Epub] CrossRef - P1 Bacteriophage-Enabled Delivery of CRISPR-Cas9 Antimicrobial Activity Against Shigella flexneri
Yang W. Huan, Vincenzo Torraca, Russell Brown, Jidapha Fa-arun, Sydney L. Miles, Diego A. Oyarzún, Serge Mostowy, Baojun Wang
ACS Synthetic Biology.2023; 12(3): 709. CrossRef - Staphylococcus aureus Dormancy: Waiting for Insurgency
Ahmad Nasser, Shiva Jahanbakhshi, Mohammad Mehdi Soltan Dallal, Maryam Banar, Azin Sattari-Maraji, Taher Azimi
Current Pharmaceutical Biotechnology.2023; 24(15): 1898. CrossRef - Recent advance on nanoparticles or nanomaterials with anti-multidrug resistant bacteria and anti-bacterial biofilm properties: A systematic review
Farhad Moradi, Arshin Ghaedi, Zahra Fooladfar, Aida Bazrgar
Heliyon.2023; 9(11): e22105. CrossRef - Isolation and characterization of lytic bacteriophages from sewage at an egyptian tertiary care hospital against methicillin-resistant Staphylococcus aureus clinical isolates
Safia Samir, Amira El-Far, Hend Okasha, Rania Mahdy, Fatima Samir, Sami Nasr
Saudi Journal of Biological Sciences.2022; 29(5): 3097. CrossRef - Staphylococcus aureus: Biofilm Formation and Strategies Against it
Ahmad Nasser , Mohammad Mehdi Soltan Dallal, Shiva Jahanbakhshi, Taher Azimi, Leila Nikouei
Current Pharmaceutical Biotechnology.2022; 23(5): 664. CrossRef - An Anti-MRSA Phage From Raw Fish Rinse: Stability Evaluation and Production Optimization
Israa M. Abd-Allah, Ghadir S. El-Housseiny, Mohammad Y. Alshahrani, Samar S. El-Masry, Khaled M. Aboshanab, Nadia A. Hassouna
Frontiers in Cellular and Infection Microbiology.2022;[Epub] CrossRef - Molecular mechanisms of Shigella effector proteins: a common pathogen among diarrheic pediatric population
Ahmad Nasser, Mehrdad Mosadegh, Taher Azimi, Aref Shariati
Molecular and Cellular Pediatrics.2022;[Epub] CrossRef - A Metal-Containing NP Approach to Treat Methicillin-Resistant Staphylococcus aureus (MRSA): Prospects and Challenges
Wendy Wai Yeng Yeo, Sathiya Maran, Amanda Shen-Yee Kong, Wan-Hee Cheng, Swee-Hua Erin Lim, Jiun-Yan Loh, Kok-Song Lai
Materials.2022; 15(17): 5802. CrossRef - Electrochemical Biosensors for Pathogen Detection: An Updated Review
Morteza Banakar, Masoud Hamidi, Zohaib Khurshid, Muhammad Sohail Zafar, Janak Sapkota, Reza Azizian, Dinesh Rokaya
Biosensors.2022; 12(11): 927. CrossRef - Isolation of a lytic bacteriophage for Helicobacter pylori
Sara Khosravi, Razieh Amini, Mohammad Reza Arabestani, Seyed Saman Talebi, Farid Azizi Jalilian
Gene Reports.2021; 23: 101107. CrossRef - Bacteriophage Therapy for Critical and High-Priority Antibiotic-Resistant Bacteria and Phage Cocktail-Antibiotic Formulation Perspective
Gursneh Kaur, Ritika Agarwal, Rakesh Kumar Sharma
Food and Environmental Virology.2021; 13(4): 433. CrossRef - A comprehensive review of bacterial osteomyelitis with emphasis on Staphylococcus aureus
Ahmad Nasser, Taher Azimi, Soheila Ostadmohammadi, Samaneh Ostadmohammadi
Microbial Pathogenesis.2020; 148: 104431. CrossRef - Characterization of the Three New Kayviruses and Their Lytic Activity Against Multidrug-Resistant Staphylococcus aureus
Natalia Łubowska, Bartłomiej Grygorcewicz, Katarzyna Kosznik-Kwaśnicka, Agata Zauszkiewicz-Pawlak, Alicja Węgrzyn, Barbara Dołęgowska, Lidia Piechowicz
Microorganisms.2019; 7(10): 471. CrossRef
- Characterization of a Vibrio parahaemolyticus-targeting lytic bacteriophage SSJ01 and its application in artificial seawater
- A Novel PCR Assay for Detecting
Brucella abortus andBrucella melitensis - Saeed Alamian, Majid Esmaelizad, Taghi Zahraei, Afshar Etemadi, Mohsen Mohammadi, Davoud Afshar, Soheila Ghaderi
- Osong Public Health Res Perspect. 2017;8(1):65-70. Published online February 28, 2017
- DOI: https://doi.org/10.24171/j.phrp.2017.8.1.09
- 4,546 View
- 66 Download
- 8 Crossref
-
 Abstract
Abstract
 PDF
PDF Objectives Brucellosis is a major zoonotic disease that poses a significant public health threat worldwide. The classical bacteriological detection process used to identify
Brucella spp. is difficult and time-consuming. This study aimed to develop a novel molecular assay for detecting brucellosis.Methods All complete sequences of chromosome 1 with 2.1-Mbp lengths were compared among all available
Brucella sequences. A unique repeat sequence (URS) locus on chromosome 1 could differentiateBrucella abortus fromBrucella melitensis . A primer set was designed to flank the unique locus. A total of 136 lymph nodes and blood samples were evaluated and classified by the URS-polymerase chain reaction (PCR) method in 2013–2014.Results Biochemical tests and bacteriophage typing as the golden standard indicated that all
Brucella spp. isolates wereB. melitensis biovar 1 andB. abortus biovar 3. The PCR results were the same as the bacteriological method for detectingBrucella spp. The sensitivity and specificity of the URS-PCR method make it suitable for detectingB. abortus andB. melitensis .Conclusion Quick detection of
B. abortus andB. melitensis can provide the most effective strategies for control of these bacteria. The advantage of this method over other presented methods is that bothB. abortus andB. melitensis are detectable in a single test tube. Furthermore, this method covered 100% of allB. melitensis andB. abortus biotypes. The development of this URS-PCR method is the first step toward the development of a novel kit for the molecular identification ofB. abortus andB. melitensis .-
Citations
Citations to this article as recorded by- Development of a simplified and cost-effective sample preparation method for genotyping of human papillomavirus by next-generation sequencing
Rungrat Jitvaropas, Ukrit Thongpoom, Vorthon Sawaswong, Kritsada Khongnomnan, Witthaya Poomipak, Kesmanee Praianantathavorn, Pornjarim Nilyanimit, Yong Poovorawan, Sunchai Payungporn
Archives of Virology.2023;[Epub] CrossRef - Bovine brucellosis – a comprehensive review
Sandip Kumar Khurana, Anju Sehrawat, Ruchi Tiwari, Minakshi Prasad, Baldev Gulati, Muhammad Zubair Shabbir, Rajesh Chhabra, Kumaragurubaran Karthik, Shailesh Kumar Patel, Mamta Pathak, Mohd. Iqbal Yatoo, Vivek Kumar Gupta, Kuldeep Dhama, Ranjit Sah, Wanpe
Veterinary Quarterly.2021; 41(1): 61. CrossRef - Survey of Zoonotic Bacterial Pathogens in Native Foxes in Central Chile: First Record of Brucella canis Exposure
Nicolás Galarce, Sebastián de la Fuente, Beatriz Escobar, Phillip Dettleff, Pedro Abalos, Juan Carlos Hormazábal, Roberto Flores, Nicole Sallaberry-Pincheira, Víctor Martínez
Animals.2021; 11(7): 1980. CrossRef - Development and validation of immunoassay for whole cell detection of Brucella abortus and Brucella melitensis
Richa Hans, Pranjal Kumar Yadav, Pushpendra Kumar Sharma, Mannan Boopathi, Duraipandian Thavaselvam
Scientific Reports.2020;[Epub] CrossRef - Laboratory Diagnostic Procedures for Human Brucellosis: An Overview of Existing Approaches
Afshar Etemadi, Rezvan Moniri, Heinrich Neubauer, Yasaman Dasteh Goli, Saeed Alamian
Jundishapur Journal of Microbiology.2019;[Epub] CrossRef - Comparison of PCR-RFLP and PFGE for determining the clonality of Brucella isolates from human and livestock specimens
Nasrin Bahmani, Reza Mirnejad, Mohammad Reza Arabestani, Parviz Mohajerie, Seyed Hamid Hashemi, Manoochehr Karami, Mohammad Yousef Alikhani
Saudi Journal of Biological Sciences.2019; 26(2): 256. CrossRef -
Designing an immunosensor for detection of
Brucella abortus
based on coloured silica nanoparticles
Arash Shams, Bahareh Rahimian Zarif, Mojtaba Salouti, Reza Shapouri, Sako Mirzaii
Artificial Cells, Nanomedicine, and Biotechnology.2019; 47(1): 2562. CrossRef - Identification of Brucella genus and eight Brucella species by Luminex bead-based suspension array
Tina S. Lusk Pfefer, Ruth Timme, Julie A. Kase
Food Microbiology.2018; 70: 113. CrossRef
- Development of a simplified and cost-effective sample preparation method for genotyping of human papillomavirus by next-generation sequencing
- Identification of
Klebsiella pneumoniae Carbapenemase-producingKlebsiella oxytoca in Clinical Isolates in Tehran Hospitals, Iran by Chromogenic Medium and Molecular Methods - Majid Validi, Mohammad Mehdi Soltan Dallal, Masoumeh Douraghi, Jalil Fallah Mehrabadi, Abbas Rahimi Foroushani
- Osong Public Health Res Perspect. 2016;7(5):301-306. Published online October 31, 2016
- DOI: https://doi.org/10.1016/j.phrp.2016.08.006
- 2,923 View
- 34 Download
- 8 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
Production of carbapenemase, especially Klebsiella pneumoniae carbapenemases (KPC), is one of the antibiotic resistance mechanisms of Enterobacteriaceae such as Klebsiella oxytoca. This study aimed to investigate and identify KPC-producing K. oxytoca isolates using molecular and phenotypic methods.
Methods
A total of 75 isolates of K. oxytoca were isolated from various clinical samples, and were verified as K. oxytoca after performing standard microbiological tests and using a polymerase chain reaction (PCR) method. An antibiotic susceptibility test was performed using a disc diffusion method according to the Clinical and Laboratory Standards Institute guidelines. CHROMagar KPC chromogenic culture media was used to examine and confirm the production of the carbapenemase enzyme in K. oxytoca isolates; in addition, PCR was used to evaluate the presence of blaKPC gene in K. oxytoca strains.
Results
Of a total of 75 K. oxytoca isolates, one multidrug resistant strain was isolated from the urine of a hospitalized woman. This strain was examined to assess its ability to produce carbapenemase enzyme; it produced a colony with a blue metallic color on the CHROMagar KPC chromogenic culture media. In addition, the blaKPC gene was confirmed by PCR. After sequencing, it was confirmed and deposited in GenBank.
Conclusion
To date, many cases of KPC-producing Enterobacteriaceae, in particular K. pneumoniae, have been reported in different countries; there are also some reports on the identification of KPC-producing K. oxytoca. Therefore, to prevent the outbreak of nosocomial infections, the early detection, control, and prevention of the spread of these strains are of great importance. -
Citations
Citations to this article as recorded by- Klebsiella oxytoca Complex: Update on Taxonomy, Antimicrobial Resistance, and Virulence
Jing Yang, Haiyan Long, Ya Hu, Yu Feng, Alan McNally, Zhiyong Zong
Clinical Microbiology Reviews.2022;[Epub] CrossRef - Development and comparison of immunochromatographic strips with four nanomaterial labels: Colloidal gold, new colloidal gold, multi-branched gold nanoflowers and Luminol-reduced Au nanoparticles for visual detection of Vibrio parahaemolyticus in seafood
Meijiao Wu, Youxue Wu, Cheng Liu, Yachen Tian, Shuiqin Fang, Hao Yang, Bin Li, Qing Liu
Aquaculture.2021; 539: 736563. CrossRef - Variation in Accessory Genes Within the Klebsiella oxytoca Species Complex Delineates Monophyletic Members and Simplifies Coherent Genotyping
Amar Cosic, Eva Leitner, Christian Petternel, Herbert Galler, Franz F. Reinthaler, Kathrin A. Herzog-Obereder, Elisabeth Tatscher, Sandra Raffl, Gebhard Feierl, Christoph Högenauer, Ellen L. Zechner, Sabine Kienesberger
Frontiers in Microbiology.2021;[Epub] CrossRef - Multidrug-Resistant Bacteria and Alternative Methods to Control Them: An Overview
Roberto Vivas, Ana Andréa Teixeira Barbosa, Silvio Santana Dolabela, Sona Jain
Microbial Drug Resistance.2019; 25(6): 890. CrossRef - Molecular typing of cytotoxin-producing Klebsiella oxytoca isolates by 16S-23S internal transcribed spacer PCR
M.M. Soltan Dallal, M. Validi, M. Douraghi, B. Bakhshi
New Microbes and New Infections.2019; 30: 100545. CrossRef - Determination of antibiotic resistance and minimum inhibitory concentration of meropenem and imipenem growth in Klebsiella strains isolated from urinary tract infection in Shahrekord educational hospitals
Farshad Kakian, Behnam Zamzad, Abolfazl Gholipour, Kiarash Zamanzad
Journal of Shahrekord University of Medical Scienc.2019; 21(2): 80. CrossRef - Evaluation the cytotoxic effect of cytotoxin-producing Klebsiella oxytoca isolates on the HEp-2 cell line by MTT assay
Mohammad Mehdi Soltan-Dallal, Majid Validi, Masoumeh Douraghi, Jalil Fallah-Mehrabadi, Leila Lormohammadi
Microbial Pathogenesis.2017; 113: 416. CrossRef - Outbreak by Hypermucoviscous Klebsiella pneumoniae ST11 Isolates with Carbapenem Resistance in a Tertiary Hospital in China
Lingling Zhan, Shanshan Wang, Yinjuan Guo, Ye Jin, Jingjing Duan, Zhihao Hao, Jingnan Lv, Xiuqin Qi, Longhua Hu, Liang Chen, Barry N. Kreiswirth, Rong Zhang, Jingye Pan, Liangxing Wang, Fangyou Yu
Frontiers in Cellular and Infection Microbiology.2017;[Epub] CrossRef
- Klebsiella oxytoca Complex: Update on Taxonomy, Antimicrobial Resistance, and Virulence
- Plasmid-Mediated Quinolone-Resistance (
qnr ) Genes in Clinical Isolates ofEscherichia coli Collected from Several Hospitals of Qazvin and Zanjan Provinces, Iran - Maryam Rezazadeh, Hamid Baghchesaraei, Amir Peymani
- Osong Public Health Res Perspect. 2016;7(5):307-312. Published online October 31, 2016
- DOI: https://doi.org/10.1016/j.phrp.2016.08.003
- 2,972 View
- 27 Download
- 29 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
Escherichia coli is regarded as the most important etiological agent of urinary tract infections. Fluoroquinolones are routinely used in the treatment of these infections; however, in recent years, a growing rate of resistance to these drugs has been reported globally. The aims of this study were to detect plasmid-mediated qnrA, qnrB, and qnrS genes among the quinolone-nonsusceptible E. coli isolates and to investigate their clonal relatedness in Qazvin and Zanjan Provinces, Iran.
Methods
A total of 200 clinical isolates of E. coli were collected from hospitalized patients. The bacterial isolates were identified through standard laboratory protocols and further confirmed using API 20E test strips. Antimicrobial susceptibility was determined by the standard disk diffusion method. Polymerase chain reaction (PCR) and sequencing were used for detecting qnrA, qnrB, and qnrS genes and the clonal relatedness of qnr-positive isolates was evaluated by enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) method.
Results
In total, 136 (68%) isolates were nonsusceptible to quinolone compounds, among which 45 (33.1%) and 71 (52.2%) isolates showed high- and low-level quinolone resistance, respectively. Of the 136 isolates, four (2.9%) isolates were positive for the qnrS1 gene. The results from ERIC-PCR revealed that two (50%) cases of qnr-positive isolates were related genetically.
Conclusion
Our study results were indicative of the presence of low frequency of qnr genes among the clinical isolates of E. coli in Qazvin and Zanjan Provinces, which emphasizes the need for establishing tactful policies associated with infection-control measures in our hospital settings. -
Citations
Citations to this article as recorded by-
Polluted surface water is a repository of antibiotic-resistant
Escherichia coli
harbouring Plasmid-Mediated Quinolone Resistance (PMQR) determinants
Abimbola Olumide Adekanmbi, Victoria Oluwanifemi Odunfa, Olabisi Comfort Akinlabi, Adedolapo Victoria Olaposi, Oreoluwa Samuel Adebowale, Zaccheaus Oluwaseun Faniran, Omowunmi Abosede Banjo
International Journal of Environmental Studies.2024; : 1. CrossRef - Phylogenetic analysis, biofilm formation, antimicrobial resistance and relationship between these characteristics in Uropathogenic Escherichia coli
Talieh Mostaghimi, Abazar Pournajaf, Ali Bijani, Mohsen Mohammadi, Mehdi Rajabnia, Mehrdad Halaji
Molecular Biology Reports.2024;[Epub] CrossRef - A review of the mechanisms that confer antibiotic resistance in pathotypes of E. coli
Sina Nasrollahian, Jay P. Graham, Mehrdad Halaji
Frontiers in Cellular and Infection Microbiology.2024;[Epub] CrossRef - Panel of primers for evaluation of antibiotic resistance genes with real time detection of results
A. S. Mokhov, A. D. Klimova, D. V. Azarov, A. E. Goncharov
Russian Journal for Personalized Medicine.2023; 3(1): 72. CrossRef - Antimicrobial resistance in bacteria isolated from peridomestic Rattus species: A scoping literature review
Theethawat Uea-Anuwong, Kaylee A. Byers, Lloyd Christian Wahl, Omid Nekouei, Yrjo Tapio Grohn, Ioannis Magouras
One Health.2023; 16: 100522. CrossRef - Prevalence and mechanisms of ciprofloxacin resistance in Escherichia coli isolated from hospitalized patients, healthy carriers, and wastewaters in Iran
Zohreh Neyestani, Farzad Khademi, Roghayeh Teimourpour, Mojtaba Amani, Mohsen Arzanlou
BMC Microbiology.2023;[Epub] CrossRef - Quinolone resistance and biofilm formation capability of uropathogenic Escherichia coli isolates from an Iranian inpatients’ population
Elham Rastegar, Yalda Malekzadegan, Reza Khashei, Nahal Hadi
Molecular Biology Reports.2023; 50(10): 8073. CrossRef - Antibiotic-resistant Escherichia coli Undergoes a Change in mcr-1 and qnr-S Expression after being Exposed to Gamma Irradiation
Ahmed G. Merdash, Gamal M. El-Sherbiny, Ahmed O. El-Gendy, Ahmed F. Azmy, Hussein M. El-Kabbany, Maged S. Ahmad
Journal of Pure and Applied Microbiology.2023; 17(4): 2620. CrossRef -
Effect of ciprofloxacin and
in vitro
gut conditions on biofilm of
Escherichia coli
isolated from clinical and environmental sources
Vankadari Aditya, Akshatha Kotian, Sreya Saikrishnan, Anusha Rohit, Divyashree Mithoor, Indrani Karunasagar, Vijaya Kumar Deekshit
Journal of Applied Microbiology.2022; 132(2): 964. CrossRef - Detection of qnrA, qnrB, and qnrS genes in Klebsiella pneumoniae and Escherichia coli isolates from leukemia patients
Mahdane Roshani, Alireza Goodarzi, Ali Hashemi, Farhad Afrasiabi, Hossein Goudarzi, Mohammadreza Arabestani
Reviews in Medical Microbiology.2022; 33(1): 14. CrossRef - Decentralized systems for the treatment of antimicrobial compounds released from hospital aquatic wastes
Manisha Sharma, Ankush Yadav, Kashyap Kumar Dubey, Joshua Tipple, Diganta Bhusan Das
Science of The Total Environment.2022; 840: 156569. CrossRef - Presence and Transfer of Antimicrobial Resistance Determinants in Escherichia coli in Pigs, Pork, and Humans in Thailand and Lao PDR Border Provinces
Chanika Pungpian, Nuananong Sinwat, Sunpetch Angkititrakul, Rangsiya Prathan, Rungtip Chuanchuen
Microbial Drug Resistance.2021; 27(4): 571. CrossRef - Symptoms, risk factors, diagnosis and treatment of urinary tract infections
Rajanbir Kaur, Rajinder Kaur
Postgraduate Medical Journal.2021; 97(1154): 803. CrossRef - Quinolone resistance (qnrA) gene in isolates of Escherichia coli collected from the Al-Hillah River in Babylon Province, Iraq
Hawraa Mohammed Al-Rafyai, Mourouge Saadi Alwash, Noor Salman Al-Khafaji
Pharmacia.2021; 68(1): 1. CrossRef - Hypervirulent and hypermucoviscous extended-spectrum β-lactamase-producing Klebsiella pneumoniae and Klebsiella variicola in Chile
F. Morales-León, A. Opazo-Capurro, C. Caro, N. Lincopan, A. Cardenas-Arias, F. Esposito, V. Illesca, M. L. Rioseco, M. Domínguez-Yévenes, C. A. Lima, H. Bello-Toledo, Gerardo González-Rocha
Virulence.2021; 12(1): 35. CrossRef - Surgical Antibiotic Prophylaxis in an Era of Antibiotic Resistance: Common Resistant Bacteria and Wider Considerations for Practice
Bradley D Menz, Esmita Charani, David L Gordon, Andrew JM Leather, S Ramani Moonesinghe, Cameron J Phillips
Infection and Drug Resistance.2021; Volume 14: 5235. CrossRef - High prevalence of plasmid-mediated quinolone resistance (PMQR) among E. coli from aquatic environments in Bangladesh
Mohammed Badrul Amin, Sumita Rani Saha, Md Rayhanul Islam, S. M. Arefeen Haider, Muhammed Iqbal Hossain, A. S. M. Homaun Kabir Chowdhury, Emily K. Rousham, Mohammad Aminul Islam, Abdelazeem Mohamed Algammal
PLOS ONE.2021; 16(12): e0261970. CrossRef -
Development of quinolone resistance and prevalence of different virulence genes among
Shigella flexneri
and
Shigella dysenteriae
in environmental water samples
B. Roy, S.K. Tousif Ahamed, B. Bandyopadhyay, N. Giri
Letters in Applied Microbiology.2020; 71(1): 86. CrossRef - Synergistic Activity of Fluoroquinolones Combining with Artesunate Against Multidrug-Resistant Escherichia coli
SiMin Wei, YueFei Yang, Wei Tian, MingJiang Liu, ShaoJie Yin, JinGui Li
Microbial Drug Resistance.2020; 26(1): 81. CrossRef - Plasmid-Mediated Quinolone Resistance in Pseudomonas aeruginosa Isolated from Burn Patients in Tehran, Iran
Azam Molapour, Amir Peymani, Parvaneh Saffarain, Narges Habibollah-Pourzereshki, Pooya Rashvand
Infectious Disorders - Drug Targets .2020; 20(1): 49. CrossRef - Plasmidic Fluoroquinolone Resistance Genes in Fluoroquinolone-Resistant and/or Extended Spectrum Beta-Lactamase-ProducingEscherichia coliStrains Isolated from Pediatric and Adult Patients Diagnosed with Urinary Tract Infection
Melisa Akgoz, Irem Akman, Asuman Begum Ates, Cem Celik, Betul Keskin, Busra Betul Ozmen Capin, Zeynep Ceren Karahan
Microbial Drug Resistance.2020; 26(11): 1334. CrossRef - Characterization of Plasmid-Mediated Quinolone Resistance and Serogroup Distributions of Uropathogenic Escherichia coli among Iranian Kidney Transplant Patients
Amin Sadeghi, Mehrdad Halaji, Amirhossein Fayyazi, Seyed Asghar Havaei, Yun Peng Chao
BioMed Research International.2020; 2020: 1. CrossRef - Antimicrobial resistance in fecal Escherichia coli isolated from poultry chicks in northern Iran
Zohreh Pourhossein, Leila Asadpour, Hadi Habibollahi, Seyedeh Tooba Shafighi
Gene Reports.2020; 21: 100926. CrossRef - Determination of antibiotic resistance genes and virulence factors in Escherichia coli isolated from Turkish patients with urinary tract infection
Azer Özad Düzgün, Funda Okumuş, Ayşegül Saral, Ayşegül Çopur Çiçek, Sedanur Cinemre
Revista da Sociedade Brasileira de Medicina Tropic.2019;[Epub] CrossRef - The emergence of antimicrobial resistance in environmental strains of the Bacteroides fragilis group
Sebastian Niestępski, Monika Harnisz, Ewa Korzeniewska, Ma. Guadalupe Aguilera-Arreola, Araceli Contreras-Rodríguez, Zofia Filipkowska, Adriana Osińska
Environment International.2019; 124: 408. CrossRef - Evaluation of Quinolone Resistance in Escherichia coli Isolates Recovered from Urine and Feces of Patients with Acute or Recurrent Urinary Tract Infection
Hossein Norouzian, Nader Shahrokhi, Shahram Sabeti, Saeid Bouzari, Mohammad Pooya
Journal of Medical Microbiology and Infectious Dis.2019; 7(4): 120. CrossRef - Plasmid-mediated quinolones resistance in clinically important bacteria
M. Omidvar Panah, M. Najafi, A. Peymani
The Journal of Qazvin University of Medical Scien.2018; 22(2): 90. CrossRef - Detection of Plasmid-Mediated qnr Genes Among the Clinical Quinolone-Resistant Escherichia coli Strains Isolated in Tehran, Iran
Reza Ranjbar, Sajjad S. Tolon, Mehrdad Sami, Reza Golmohammadi
The Open Microbiology Journal.2018; 12(1): 248. CrossRef - Characterization of Escherichia coli strains isolated from raw vegetables
Pavel Pleva, Magda Janalíková, Silvie Pavlíčková, Martin Lecomte, Tanguy Godillon, Ivan Holko
Potravinarstvo Slovak Journal of Food Sciences.2018; 12(1): 304. CrossRef
-
Polluted surface water is a repository of antibiotic-resistant
Escherichia coli
harbouring Plasmid-Mediated Quinolone Resistance (PMQR) determinants
- One-Step Reverse Transcription-Polymerase Chain Reaction for Ebola and Marburg Viruses
- Sun-Whan Park, Ye-Ji Lee, Won-Ja Lee, Youngmee Jee, WooYoung Choi
- Osong Public Health Res Perspect. 2016;7(3):205-209. Published online June 30, 2016
- DOI: https://doi.org/10.1016/j.phrp.2016.04.004
- 2,810 View
- 23 Download
- 5 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
Ebola and Marburg viruses (EBOVs and MARVs, respectively) are causative agents of severe hemorrhagic fever with high mortality rates in humans and nonhuman primates. In 2014, there was a major Ebola outbreak in various countries in West Africa, including Guinea, Liberia, Republic of Sierra Leone, and Nigeria. EBOV and MARV are clinically difficult to diagnose and distinguish from other African epidemic diseases. Therefore, in this study, we aimed to develop a method for rapid identification of the virus to prevent the spread of infection.
Methods
We established a conventional one-step reverse transcription-polymerase chain reaction (RT-PCR) assay for these pathogens based on the Superscript Reverse Transcriptase-Platinum Taq polymerase enzyme mixture. All assays were thoroughly optimized using in vitro-transcribed RNA.
Results
We designed seven primer sets of nucleocapsid protein (NP) genes based on sequences from seven filoviruses, including five EBOVs and two MARVs. To evaluate the sensitivity of the RT-PCR assay for each filovirus, 10-fold serial dilutions of synthetic viral RNA transcripts of EBOV or MARV NP genes were used to assess detection limits of viral RNA copies. The potential for these primers to cross react with other filoviruses was also examined. The results showed that the primers were specific for individual genotype detection in the examined filoviruses.
Conclusion
The assay established in this study may facilitate rapid, reliable laboratory diagnosis in suspected cases of Ebola and Marburg hemorrhagic fevers. -
Citations
Citations to this article as recorded by- Marburg Virus Disease – A Mini-Review
Sandip Chakraborty, Deepak Chandran, Ranjan K. Mohapatra, Mahmoud Alagawany, Mohd Iqbal Yatoo, Md. Aminul Islam, Anil K. Sharma, Kuldeep Dhama
Journal of Experimental Biology and Agricultural S.2022; 10(4): 689. CrossRef - Marburgviruses: An Update
Caterina M Miraglia
Laboratory Medicine.2019; 50(1): 16. CrossRef - Ebola virus: A global public health menace: A narrative review
Shamimul Hasan, SyedAnsar Ahmad, Rahnuma Masood, Shazina Saeed
Journal of Family Medicine and Primary Care.2019; 8(7): 2189. CrossRef - Fast and Parallel Detection of Four Ebola Virus Species on a Microfluidic-Chip-Based Portable Reverse Transcription Loop-Mediated Isothermal Amplification System
Xue Lin, Xiangyu Jin, Bin Xu, Ruliang Wang, Rongxin Fu, Ya Su, Kai Jiang, Han Yang, Ying Lu, Yong Guo, Guoliang Huang
Micromachines.2019; 10(11): 777. CrossRef - The current landscape of nucleic acid tests for filovirus detection
David J. Clark, John Tyson, Andrew D. Sails, Sanjeev Krishna, Henry M. Staines
Journal of Clinical Virology.2018; 103: 27. CrossRef
- Marburg Virus Disease – A Mini-Review
- Comparison of Three Different Methods for Detection of IL28 rs12979860 Polymorphisms as a Predictor of Treatment Outcome in Patients with Hepatitis C Virus
- Abolfazl Fateh, Mohammadreza Aghasadeghi, Seyed D. Siadat, Farzam Vaziri, Farzin Sadeghi, Roohollah Fateh, Hossein Keyvani, Alireza H. Tasbiti, Shamsi Yari, Angila Ataei-Pirkooh, Seyed H. Monavari
- Osong Public Health Res Perspect. 2016;7(2):83-89. Published online April 30, 2016
- DOI: https://doi.org/10.1016/j.phrp.2015.11.004
- 3,037 View
- 19 Download
- 22 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
This study aimed to evaluate the specificity, sensitivity, cost, and turn-around time of three methods of gene polymorphism analysis and to study the relationship between IL28B rs12979860 and SVR rate to pegIFN-α/RVB therapy among patients with chronic hepatitis C.
Methods
A total of 100 samples from chronic hepatitis C patients were analyzed in parallel using the three methods: direct sequencing, real-time polymerase chain reaction (PCR), amplification refractory mutation system (ARMS)-PCR.
Results
The different profiles for IL28B rs12979860 alleles (CC, CT, and TT) obtained with PCR-RFLP, ARMS-PCR, and direct sequencing were consistent among the three methods. Prevalence of rs12979860 genotypes CC, CT and TT in HCV genotype 1a was 10(19.6%), 35(68.6%), and six (11.8%), respectively, and in HCV genotype 31, it was 13(26.5%), 31(63.3%), and five (10.2%), respectively. No significant difference was seen between rs12979860 genotype and HCV genotype (p = 0.710).
Conclusion
Screening by ARMS – PCR SNOP detection represents the most efficient and reliable method to determine HCV polymorphisms in routine clinical practice. -
Citations
Citations to this article as recorded by- Multiplex Snapshot Minisequencing for the Detection of Common PAH Gene Mutations in Iranian Patients with Phenylketonuria
Pegah Namdar Aligoodarzi, Golale Rostami, Seyed Reza Kazemi Nezhad, Mohammad Hamid
Iranian Biomedical Journal.2023; 27(1): 46. CrossRef - Expression of TRIM56 gene in SARS-CoV-2 variants and its relationship with progression of COVID-19
Rezvan Tavakoli, Pooneh Rahimi, Mojtaba Hamidi-Fard, Sana Eybpoosh, Delaram Doroud, Iraj Ahmadi, Enayat Anvari, Mohammadreza Aghasadeghi, Abolfazl Fateh
Future Virology.2023; 18(9): 563. CrossRef - Performance of the tetra-primer PCR technique compared to PCR-RFLP in the search for rs12979860 (C/T) and rs8099917 (T/G) single nucleotide polymorphisms (SNPs) in the IFNL4 gene
Ellen Hochleitner Souza Kindermann, Karoline Rodrigues Campos, Adele Caterino-de-Araujo
Revista do Instituto Adolfo Lutz.2023; 82: 1. CrossRef - Glioblastoma as a Novel Drug Repositioning Target: Updated State
Hamed Hosseinalizadeh, Ammar Ebrahimi, Ahmad Tavakoli, Seyed Hamidreza Monavari
Anti-Cancer Agents in Medicinal Chemistry.2023; 23(11): 1253. CrossRef - Prevalence of Human Cytomegalovirus Infection in Iranian Prostate Cancer Patients
Ehsan Alborzi, Ahmad Tavakoli, Seyed Jalal Kiani, Saied Ghorbani, Davod Javanmard, Milad Sabaei, Maryam Fatemipour, Seyed Hamidreza Monavari
Iranian Journal of Medical Microbiology.2023; 17(4): 379. CrossRef - MicroRNAs Profiling in HIV, HCV, and HIV/HCV Co-Infected Patients
Mohsen Moghoofei, Sohrab Najafipour, Shayan Mostafaei , Ahmad Tavakoli , Farah Bokharaei-Salim , Saied Ghorbani, Davod Javanmard, Hadi Ghaffari , Seyed Hamidreza Monavari
Current HIV Research.2021; 19(1): 27. CrossRef - Occult hepatitis C virus infection in hemophilia patients and its correlation with interferon lambda 3 and 4 polymorphisms
Amir Hossein Nafari, Ahmad Ayadi, Zahra Noormohamadi, Fatemeh Sakhaee, Farzam Vaziri, Seyed Davar Siadat, Abolfazl Fateh
Infection, Genetics and Evolution.2020; 79: 104144. CrossRef - Polymerase Chain Reaction Assay Using the Restriction Fragment Length Polymorphism Technique in the Detection of Prosthetic Joint Infections: A Multi-Centered Study
Ataollah Moshirabadi, Mohammad Razi, Peyman Arasteh, Mohammad Mahdi Sarzaeem, Saman Ghaffari, Saied Aminiafshar, Kami Hosseinian Khosroshahy, Fatemeh Maryam Sheikholeslami
The Journal of Arthroplasty.2019; 34(2): 359. CrossRef - One-Step ARMS-PCR for the Detection of SNPs—Using the Example of the PADI4 Gene
Ehnert, Linnemann, Braun, Botsch, Leibiger, Hemmann, Nussler
Methods and Protocols.2019; 2(3): 63. CrossRef - Epstein–Barr virus and risk of breast cancer: a systematic review and meta-analysis
Mohammad Farahmand, Seyed Hamidreza Monavari, Zabihollah Shoja, Hadi Ghaffari, Mehdi Tavakoli, Ahmad Tavakoli
Future Oncology.2019; 15(24): 2873. CrossRef - Modeling suggests that microliter volumes of contaminated blood caused an outbreak of hepatitis C during computerized tomography
Eyal Shteyer, Louis Shekhtman, Tal Zinger, Sheri Harari, Inna Gafanovich, Dana Wolf, Hefziba Ivgi, Rima Barsuk, Ilana Dery, Daniela Armoni, Mila Rivkin, Rahul Pipalia, Michal Cohen Eliav, Yizhak Skorochod, Gabriel S. Breuer, Ran Tur-kaspa, Yonit Weil Wien
PLOS ONE.2019; 14(1): e0210173. CrossRef - Correlation of CD81 and SCARB1 polymorphisms on virological responses in Iranian patients with chronic hepatitis C virus genotype 1
Milad Nafari, Shiva Irani, Farzam Vaziri, Safoora Gharibzadeh, Fatemeh Sakhaee, Mohammad Khazeni, Naser Kalhor, Fatemeh Rahimi Jamnani, Seyed Davar Siadat, Abolfazl Fateh
Infection, Genetics and Evolution.2018; 62: 296. CrossRef - First detection of human hepegivirus-1 (HHpgV-1) in Iranian patients with hemophilia
Yazdan Bijvand, Mohammad Reza Aghasadeghi, Fatemeh Sakhaee, Parviz Pakzad, Farzam Vaziri, Alireza Azizi Saraji, Fatemeh Rahimi Jamnani, Seyed Davar Siadat, Abolfazl Fateh
Scientific Reports.2018;[Epub] CrossRef - Evaluation of TRIM5 and TRIM22 polymorphisms on treatment responses in Iranian patients with chronic hepatitis C virus infection
Setareh Mobasheri, Nazanin Irani, Abbas Akhavan Sepahi, Fatemeh Sakhaee, Fatemeh Rahimi Jamnani, Farzam Vaziri, Seyed Davar Siadat, Abolfazl Fateh
Gene.2018; 676: 95. CrossRef - Data Mining and Machine Learning Algorithms Using IL28B Genotype and Biochemical Markers Best Predicted Advanced Liver Fibrosis in Chronic Hepatitis C
Hend Ibrahim Shousha, Abubakr Hussein Awad, Dalia Abdelhamid Omran, Mayada Mohamed Elnegouly, Mahasen Mabrouk
Japanese Journal of Infectious Diseases.2018; 71(1): 51. CrossRef - IL28B rs12980275 and HLA rs4273729 genotypes as a powerful predictor factor for rapid, early, and sustained virologic response in patients with chronic hepatitis C
Parvaneh Sedighimehr, Shiva Irani, Fatemeh Sakhaee, Farzam Vaziri, Mohammadreza Aghasadeghi, Seyed Mehdi Sadat, Fatemeh Rahimi Jamnani, Abolfazl Fateh, Seyed Davar Siadat
Archives of Virology.2017; 162(1): 181. CrossRef - Comparison of Direct Sequencing, Real-Time PCR-High Resolution Melt (PCR-HRM) and PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) Analysis for Genotyping of Common Thiopurine Intolerant Variant Alleles NUDT15 c.415C>T and TPMT c.719A>G (TPMT*3C)
Wai-Ying Fong, Chi-Chun Ho, Wing-Tat Poon
Diagnostics.2017; 7(2): 27. CrossRef - Effect of IL15 rs10833 and SCARB1 rs10846744 on virologic responses in chronic hepatitis C patients treated with pegylated interferon-α and ribavirin
Sahar Sadeghi, Mehdi Davari, Esmaeil Asli, Safoora Gharibzadeh, Farzam Vaziri, Fatemeh Rahimi Jamnani, Abolfazl Fateh, Seyed Davar Siadat
Gene.2017; 630: 28. CrossRef - High platelet count and high probability of CALR detection in myeloproliferative neoplasms
Reza Shirzad, Zari Tahan-nejad, Javad Mohamadi-asl, Mohammad Seghatoleslami, Ahmad Ahmadzadeh, Amal Saki Malehi, Najmaldin Saki
Comparative Clinical Pathology.2017; 26(1): 25. CrossRef - A comparative study of various methods for detection of IL28B rs12979860 in chronic hepatitis C
Seyed Hamidreza Monavari, Roohollah Fateh, Farzam Vaziri, Fatemeh Rahimi Jamnani, Enayat Anvari, Farzin Sadeghi, Parviz Afrough, Ava Behrouzi, Fatemeh Sakhaee, Sepideh Meidaninikjeh, Hamidreza Mollaie, Alireza Hadizadeh Tasbiti, Shamsi Yari, Maryam Sadegh
Scandinavian Journal of Clinical and Laboratory In.2017; 77(4): 247. CrossRef - EGFR rs11506105 and IFNL3 SNPs but not rs8099917 are strongly associated with treatment responses in Iranian patients with chronic hepatitis C
M Asnavandi, M Zargar, F Vaziri, F R Jamnani, S Gharibzadeh, A Fateh, S D Siadat
Genes & Immunity.2017; 18(3): 144. CrossRef - Genetic Variation in Interleukin-28B and Response to Peg-IFNα-2a/RBV Combination Therapy in Patients with Hepatitis C Virus Infection
Farah Bokharaei-Salim, Mostafa Salehi-Vaziri, Farzin Sadeghi, Khadijeh Khanaliha, Maryam Esghaei, Seyed Hamidreza Monavari, Seyed Moayed Alavian, Shahin Fakhim, Hossein Keyvani
Jundishapur Journal of Microbiology.2016;[Epub] CrossRef
- Multiplex Snapshot Minisequencing for the Detection of Common PAH Gene Mutations in Iranian Patients with Phenylketonuria
- Study on the Behavior of Dengue Viruses during Outbreaks with Reference to Entomological and Laboratory Surveillance in the Cuddalore, Nagapattinam, and Tirunelveli Districts of Tamil Nadu, India
- Parasuraman Basker, Karumana Gounder Kolandaswamy
- Osong Public Health Res Perspect. 2015;6(3):143-158. Published online June 30, 2015
- DOI: https://doi.org/10.1016/j.phrp.2015.05.001
- 2,840 View
- 26 Download
- 6 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
This study was carried out in order to understand the behavior of dengue viruses through the entomological and laboratory surveillance of outbreaks. The aim of the study was to provide additional research to support current knowledge of epidemiological, clinical, and laboratory diagnosis of dengue virus and ultimately to use this information to forecast dengue as well as to justify intervention measures.
Methods
Data on the presence of Aedes larvae in human dwellings during the entomological surveillance in Cuddalore, Nagapattinam, and Tirunelveli dengue outbreaks were taken to compute indices, namely the House Index (HI), Container index (CI), and the Breteau Index (BI). Standard procedures were followed for nonstructural Protein 1 (NS1) and immunoglobulin M enzyme linked immunosorbent assay for the confirmation of dengue. Serovar confirmation was made in the Kottayam field station of the Vector Control Research Center, Puducherry.
Results
Larval indices HI < 2–3% and BI < 20 contributed to halting the outbreak. Incubation of the dengue viruses in humans was detected at 15 days, NS1 was identified as a tool for the early diagnosis of dengue cases and its presence indicated the need to implement all available interventions. It was also discovered that it is helpful to search for hidden habitats of Aedes when dengue cases have not been reduced even after the sustainable management of the larval indices, HI < 5% and BI < 20. Based on the observed incidences of stopping dengue outbreaks, it was learnt that neighborhood areas of the outbreak villages, around 400 m, should have permissible larval indices < 5% HI and BI < 20. Heterogeneous serovars that led to dengue hemorrhagic fever and Dengue Shock Syndrome (DSS) were identified using reverse transcription polymerase chain reaction and reconfirmed in the field as DEN-1 and DEN-3 viruses and were circulating in Tirunelveli during the outbreak.
Conclusion
The behaviors of dengue viruses experienced in experimental, clinical, epidemiological, entomological, and laboratory surveillance did not deviate from observations in the field during dengue outbreaks in the Cuddalore, Nagapattinam, and Tirunelveli districts of Tamil Nadu, India. -
Citations
Citations to this article as recorded by- A systematic review of published literature on mosquito control action thresholds across the world
Vindhya S. Aryaprema, Madeline R. Steck, Steven T. Peper, Rui-de Xue, Whitney A. Qualls, Olaf Horstick
PLOS Neglected Tropical Diseases.2023; 17(3): e0011173. CrossRef - Application of Nanomaterials in the Field of New Energy Environment and Economic Benefit Analysis
Shuai Xu, Haoying An, Jiahai Dai, Haichang Zhang
Advances in Materials Science and Engineering.2022; 2022: 1. CrossRef - A study on stegomyia indices in dengue control: a fuzzy approach
Sayani Adak, Soovoojeet Jana
Soft Computing.2021; 25(1): 699. CrossRef - Epidemiology and challenges of dengue surveillance in the WHO South-East Asia Region
Tsheten Tsheten, Darren J Gray, Archie C A Clements, Kinley Wangdi
Transactions of The Royal Society of Tropical Medi.2021; 115(6): 583. CrossRef - Assessing the interplay between dengue incidence and weather in Jakarta via a clustering integrated multiple regression model
Muhammad Fakhruddin, Prama Setia Putra, Karunia Putra Wijaya, Ardhasena Sopaheluwakan, Ratna Satyaningsih, Kurnia Endah Komalasari, Mamenun, Sumiati, Sapto Wahyu Indratno, Nuning Nuraini, Thomas Götz, Edy Soewono
Ecological Complexity.2019; 39: 100768. CrossRef - Emergence of Dengue Virus 4 as the Predominant Serotype during the Outbreak of 2017 in South India
P.Ferdinamarie Sharmila, K. Vanathy, Barathidasan Rajamani, Venkatesh Kaliaperumal, Rahul Dhodapkar
Indian Journal of Medical Microbiology.2019; 37(3): 393. CrossRef
- A systematic review of published literature on mosquito control action thresholds across the world
- Evaluation and Comparison of Molecular and Conventional Diagnostic Tests for Detecting Tuberculosis in Korea, 2013
- Sang-Hee Park, Chang-Ki Kim, Hye-Ran Jeong, Hyunjin Son, Seong-Han Kim, Mi-Sun Park
- Osong Public Health Res Perspect. 2014;5(Suppl):S3-S7. Published online December 31, 2014
- DOI: https://doi.org/10.1016/j.phrp.2014.10.006
- 2,898 View
- 20 Download
- 7 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
A fast and accurate diagnosis is necessary to control and eliminate tuberculosis (TB). In Korea, TB continues to be a serious public health problem. In this study, diagnostic tests on clinical samples from patients suspected to have TB were performed and the sensitivity and specificity of the various techniques were compared. The main objective of the study was to compare various diagnostic tests and evaluate their sensitivity and specificity for detecting tuberculosis.
Methods
From January 2013 to December 2013, 170,240 clinical samples from patients suspected to have TB were tested with smear microscopy, acid-fast bacilli culture, and real-time polymerase chain reaction (PCR). The test results were compared and data were analyzed.
Results
A total of 8216 cultures tested positive for TB (positive detection rate, 4.8%). The contamination rate in the culture was 0.6% and the isolation rate of nontuberculous mycobacteria was 1.0%. The sensitivity and specificity of smear microscopy were 56.8% and 99.6%, respectively. The concordance rate between the solid and liquid cultures was 92.8%. Mycobacterium isolates were not detected in 0.4% of the cases in the liquid culture, whereas no Mycobacterium isolates were detected in 6.8% of the cases in the solid culture. The sensitivity and specificity of real-time PCR for the solid culture were 97.2% and 72.4%, respectively, whereas the corresponding data for the liquid culture were 93.5% and 97.2%.
Conclusion
The study results can be used to improve existing TB diagnosis procedure as well as for comparing the effectiveness of the assay tests used for detecting Mycobacterium tuberculosis isolates. -
Citations
Citations to this article as recorded by- The Xpert® MTB/RIF diagnostic test for pulmonary and extrapulmonary tuberculosis in immunocompetent and immunocompromised patients: Benefits and experiences over 2 years in different clinical contexts
Ana Paula de Oliveira Tomaz, Sonia Mara Raboni, Gislene Maria Botão Kussen, Keite da Silva Nogueira, Clea Elisa Lopes Ribeiro, Libera Maria Dalla Costa, Padmapriya P. Banada
PLOS ONE.2021; 16(3): e0247185. CrossRef - Comparative performance of the laboratory assays used by a Diagnostic Laboratory Hub for opportunistic infections in people living with HIV
Narda Medina, Ana Alastruey-Izquierdo, Danicela Mercado, Oscar Bonilla, Juan C. Pérez, Luis Aguirre, Blanca Samayoa, Eduardo Arathoon, David W. Denning, Juan Luis Rodriguez-Tudela
AIDS.2020; 34(11): 1625. CrossRef - Multiplex PCR is a Rapid, Simple and Cheap Method for Direct Diagnosis of M. tuberculosis from Sputum Samples
Tarig M.S. Alnour, Faisel Abuduhier , Mohammed Khatatneh , Fahad Albalawi , Khalid Alfifi , Bernard Silvala
Infectious Disorders - Drug Targets .2020; 20(4): 495. CrossRef - Naked eye detection of the Mycobacterium tuberculosis complex by recombinase polymerase amplification—SYBR green I assays
Nuntita Singpanomchai, Yukihiro Akeda, Kazunori Tomono, Aki Tamaru, Pitak Santanirand, Panan Ratthawongjirakul
Journal of Clinical Laboratory Analysis.2019;[Epub] CrossRef - Retropharyngeal SOL: An unusual presentation of a multifaceted entity
Poojan Agarwal, Manju Kaushal, Shruti Dogra, Ankur Gupta, Nishi Sharma
CytoJournal.2018; 15: 12. CrossRef - Rapid Detection of Rifampicin- and Isoniazid-Resistant Mycobacterium tuberculosis using TaqMan Allelic Discrimination
Davood Darban-Sarokhalil, Mohammad J. Nasiri, Abbas A.I. Fooladi, Parvin Heidarieh, Mohammad M. Feizabadi
Osong Public Health and Research Perspectives.2016; 7(2): 127. CrossRef - Port site infection in laparoscopic surgery: A review of its management
Prakash K Sasmal
World Journal of Clinical Cases.2015; 3(10): 864. CrossRef
- The Xpert® MTB/RIF diagnostic test for pulmonary and extrapulmonary tuberculosis in immunocompetent and immunocompromised patients: Benefits and experiences over 2 years in different clinical contexts
- Acute Human Cytomegalovirus Infection with Bleeding in Iran
- Behzad Pourhossein, Farhad Yaghmaei, Saber Esmaeili, Omid Banafshi, Shahla Afrasiabian, Mohammad Reza Shirzadi, Mark Schleiss, Ehsan Mostafavi
- Osong Public Health Res Perspect. 2014;5(6):383-386. Published online December 31, 2014
- DOI: https://doi.org/10.1016/j.phrp.2014.10.003
- 2,521 View
- 27 Download
-
 Abstract
Abstract
 PDF
PDF - In December 2011, a 42-year-old male farmer was admitted to a hospital in Sanandaj (Western Iran) with fever and anemia in order to check whether he suffered from some infectious diseases. During the first 3 days after admission, the patient gradually developed progressive oliguria, fever, abdominal pain in the right upper quadrant, leukocytosis with toxic granulation, petechiae and ecchymosis, oral bleeding, and vomiting. The sonographic findings revealed splenomegaly and an increase in the thickness of the gall bladder wall. In order to manage the patient and taking into consideration the most probable differential diagnoses, diagnostic tests were performed on two blood samples collected from him, and real-time polymerase chain reaction for human cytomegalovirus was positive.
- Molecular Investigation of Quinolone Resistance of Quinolone Resistance-Determining Region in
Streptococcus pneumoniae Strains Isolated from Iran Using Polymerase Chain Reaction–Restriction Fragment Length Polymorphism Method - Mohammad Kargar, Fataneh Moein Jahromi, Abbas Doosti, Somayeh Handali
- Osong Public Health Res Perspect. 2014;5(5):245-250. Published online October 31, 2014
- DOI: https://doi.org/10.1016/j.phrp.2014.08.010
- 2,847 View
- 22 Download
- 2 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
The resistance of Streptococcus pneumoniae to the recently available antibiotic treatment has been a growing problem. The aim of the study was to determine the quinolone-resistant strains and detect the presence of mutations in the quinolone resistance-determining regions of the gyrA, parE, and parC genes.
Methods
In this study, for the first time in Iran, the polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) method was used to investigate the presence of mutations at quinolone resistance-determining regions of topoisomerase IV and DNA gyrase on 82 S. pneumoniae strains, among them 45 clinical samples were from patients and 37 from healthy carriers (control group).
Results
In clinical samples, 34 (75.56%) strains contained mutations in the parC gene, 31 (68.89%) carried mutations in the gyrA gene, and 14 (31.11%) had parE gene mutations. Antibiotic susceptibility test was performed using the CLSI (Clinical and Laboratory Standards Institute) criteria on three different generations of quinolone family, with nalidixic acid (82.22%) showing the highest resistance and levofloxacin (42.22%) the least resistance.
Conclusion
Results
indicated that there is a significant correlation between quinolone resistance development and mutations in the parE gene as well as in the parC and gyrA genes. -
Citations
Citations to this article as recorded by- DNA-based detection of pork content in food
Muflihah, Ari Hardianto, Pintaka Kusumaningtyas, Sulistyo Prabowo, Yeni Wahyuni Hartati
Heliyon.2023; 9(3): e14418. CrossRef - Mutant selection window of four quinolone antibiotics against clinical isolates of Streptococcus pneumoniae , Haemophilus influenzae and Moraxella catarrhalis
Hajime Nakai, Takumi Sato, Takashi Uno, Emiko Furukawa, Masato Kawamura, Hiroshi Takahashi, Akira Watanabe, Shigeru Fujimura
Journal of Infection and Chemotherapy.2018; 24(2): 83. CrossRef
- DNA-based detection of pork content in food
- Travel-Associated Chikungunya Cases in South Korea during 2009–2010
- Go Woon Cha, Jung Eun Cho, Eun Ju Lee, Young Ran Ju, Myung Guk Han, Chan Park, Young Eui Jeong
- Osong Public Health Res Perspect. 2013;4(3):170-175. Published online June 30, 2013
- DOI: https://doi.org/10.1016/j.phrp.2013.04.008
- 3,027 View
- 14 Download
- 10 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
Chikungunya (CHIK) has been classified as a communicable disease group IV in South Korea since late 2010. Based on this, we investigated the extent of imported cases of CHIK in dengue-suspected individuals returning from dengue-endemic regions.
Methods
A total of 486 dengue-suspected serum samples were screened for CHIK by enzyme-linked immunosorbent assay (ELISA) and reverse transcription-polymerase chain reaction (RT-PCR) analysis. Further RT-PCR-positive samples were used for the viral culture, and CHIK was subsequently confirmed by sequence analysis of the culture samples.
Results
Five out of 107 dengue-positive samples were found to be positive for CHIK and 15 out of 379 dengue-negative samples were found to be positive for CHIK by immunoglobulin M ELISA. Further, a CHIK virus was isolated from one of the two RT-PCR-positive sera by cell culture and confirmed by sequence analysis.
Conclusion
The present study documents the first evidence of travel-associated CHIK infection in South Korea. Considering the intense international traffic between countries, our finding emphasizes the urgent need for active patient and vector surveillance for timely response to reduce the introduction of CHIK in Korea. -
Citations
Citations to this article as recorded by- Global prevalence of dengue and chikungunya coinfection: A systematic review and meta-analysis of 43,341 participants
Ahmad Adebayo Irekeola, E.A. R Engku Nur Syafirah, Md Asiful Islam, Rafidah Hanim Shueb
Acta Tropica.2022; 231: 106408. CrossRef - Current Status and a Perspective of Mosquito-Borne Diseases in the Republic of Korea
Jae Hyoung Im, Tong-Soo Kim, Moon-Hyun Chung, Ji Hyeon Baek, Hea Yoon Kwon, Jin-Soo Lee
Vector-Borne and Zoonotic Diseases.2021; 21(2): 69. CrossRef - Implications of a travel connectivity-based approach for infectious disease transmission risks in Oceania
Angela Cadavid Restrepo, Luis Furuya-Kanamori, Helen Mayfield, Eric Nilles, Colleen L Lau
BMJ Open.2021; 11(8): e046206. CrossRef - Development of a neutralization assay based on the pseudotyped chikungunya virus of a Korean isolate
Woo-Chang Chung, Kwang Yeon Hwang, Suk-Jo Kang, Jae-Ouk Kim, Moon Jung Song
Journal of Microbiology.2020; 58(1): 46. CrossRef - Chikungunya virus infection in Indonesia: a systematic review and evolutionary analysis
Harapan Harapan, Alice Michie, Mudatsir Mudatsir, Roy Nusa, Benediktus Yohan, Abram Luther Wagner, R. Tedjo Sasmono, Allison Imrie
BMC Infectious Diseases.2019;[Epub] CrossRef - Global prevalence and distribution of coinfection of malaria, dengue and chikungunya: a systematic review
Nasir Salam, Shoeb Mustafa, Abdul Hafiz, Anis Ahmad Chaudhary, Farah Deeba, Shama Parveen
BMC Public Health.2018;[Epub] CrossRef - Chikungunya Virus Infection after Traveling to Surinam, South America
Hoe-Soo Jang, Jong-Hun Chung, Joa Kim, Sun Ae Han, Na-Ra Yun, Dong-Min Kim
The Korean Journal of Medicine.2016; 90(3): 262. CrossRef - Molecular epidemiology, evolution and phylogeny of Chikungunya virus: An updating review
Alessandra Lo Presti, Eleonora Cella, Silvia Angeletti, Massimo Ciccozzi
Infection, Genetics and Evolution.2016; 41: 270. CrossRef - The First Imported Case Infected with Chikungunya Virus in Korea
Jeong-Hwan Hwang, Chang-Seop Lee
Infection & Chemotherapy.2015; 47(1): 55. CrossRef - Zika virus in Brazil and the danger of infestation by Aedes (Stegomyia) mosquitoes
Carlos Brisola Marcondes, Maria de Fátima Freire de Melo Ximenes
Revista da Sociedade Brasileira de Medicina Tropic.2015; 49(1): 4. CrossRef
- Global prevalence of dengue and chikungunya coinfection: A systematic review and meta-analysis of 43,341 participants
- Identification of Dengue Type 1 Virus (DENV-1) in Koreans Traveling Abroad
- Young Eui Jeong, Yeon Hee Kim, Jung Eun Cho, Myung Guk Han, Young Ran Ju
- Osong Public Health Res Perspect. 2011;2(1):34-40. Published online June 30, 2011
- DOI: https://doi.org/10.1016/j.phrp.2011.04.002
- 2,833 View
- 15 Download
- 15 Crossref
-
 Abstract
Abstract
 PDF
PDF - Objectives
To date, no indigenous dengue virus (DENV) transmissions have been reported in Korea. However, imported dengue infections have been diagnosed in travelers returning from endemic areas. This study presents the first virological evidence of travel-associated DENV importation into South Korea.
Methods
From January 2004 to June 2006, a total of 278 serum samples from 245 patients with suspected dengue fever were tested using the Panbio Dengue Duo IgM/IgG Rapid Strip Test. We selected 11 of the early symptomatic-phase sera that were negative for IgM and retrospectively studied them by virus isolation and reverse transcription-polymerase chain reaction.
Results
All 11 serum samples were found to be DENV positive by reverse transcription-polymerase chain reaction and viruses were successfully isolated from seven of the 11 serum samples. All the isolates were identified as DENV serotype-1.
Conclusion
We successfully isolated seven DENV serotype-1 strains for the first time in South Korea from imported infections. Considering that the vector mosquito, Aedes albopictus, already exists in South Korea, we propose that a vector surveillance program for dengue is urgently needed. -
Citations
Citations to this article as recorded by- Genotypic persistence of dengue virus in the Philippines
Ma. Jowina Galarion, Brian Schwem, Coleen Pangilinan, Angelo dela Tonga, Joy Ann Petronio-Santos, Erlinda delos Reyes, Raul Destura
Infection, Genetics and Evolution.2019; 69: 134. CrossRef - Neutralization of Acidic Intracellular Vesicles by Niclosamide Inhibits Multiple Steps of the Dengue Virus Life Cycle In Vitro
Eunhye Jung, Sangwoo Nam, Hyeryeon Oh, Sangmi Jun, Hyun-Joo Ro, Baek Kim, Meehyein Kim, Yun Young Go
Scientific Reports.2019;[Epub] CrossRef - Seroprevalence of Dengue Virus Antibody in Korea
Ji Hyen Lee, Han Wool Kim, Kyung-Hyo Kim
Pediatric Infection & Vaccine.2018; 25(3): 132. CrossRef - Circulating Genotypes of Dengue-1 Virus in South West India, 2014–2015
Shetty Pooja, Sasidharanpillai Sabeena, Bhaskar Revti, Ramachandran Sanjay, Aithal Anjali, Kumar Rajendra, Sushama Aswathyraj, Dsouza Giselle, Maity Hindol, Govindakarnavar Arunkumar
Japanese Journal of Infectious Diseases.2017; 70(6): 663. CrossRef - WITHDRAWN: A disease around the corner
Hae-Wol Cho, Chaeshin Chu
Osong Public Health and Research Perspectives.2016;[Epub] CrossRef - A Disease Around the Corner
Hae-Wol Cho, Chaeshin Chu
Osong Public Health and Research Perspectives.2016; 7(1): 1. CrossRef - Prevalence of Neutralizing Antibodies to Japanese Encephalitis Virus among High-Risk Age Groups in South Korea, 2010
Eun Ju Lee, Go-Woon Cha, Young Ran Ju, Myung Guk Han, Won-Ja Lee, Young Eui Jeong, Nagendra R Hegde
PLOS ONE.2016; 11(1): e0147841. CrossRef - Laboratory-acquired dengue virus infection by needlestick injury: a case report, South Korea, 2014
Changhwan Lee, Eun Jung Jang, Donghyok Kwon, Heun Choi, Jung Wan Park, Geun-Ryang Bae
Annals of Occupational and Environmental Medicine.2016;[Epub] CrossRef - A Pan-Dengue Virus Reverse Transcription-Insulated Isothermal PCR Assay Intended for Point-of-Need Diagnosis of Dengue Virus Infection by Use of the POCKIT Nucleic Acid Analyzer
Yun Young Go, R. P. V. Jayanthe Rajapakse, Senanayake A. M. Kularatne, Pei-Yu Alison Lee, Keun Bon Ku, Sangwoo Nam, Pin-Hsing Chou, Yun-Long Tsai, Yu-Lun Liu, Hsiao-Fen Grace Chang, Hwa-Tang Thomas Wang, Udeni B. R. Balasuriya, M. J. Loeffelholz
Journal of Clinical Microbiology.2016; 54(6): 1528. CrossRef - Comparison of the Epidemiological Aspects of Imported Dengue Cases between Korea and Japan, 2006–2010
Young Eui Jeong, Won-Chang Lee, Jung Eun Cho, Myung-Guk Han, Won-Ja Lee
Osong Public Health and Research Perspectives.2016; 7(1): 71. CrossRef - Complete Genome Sequences of Three Clinical Isolates of Dengue Virus Serotype 1 from South Korean Travelers
Yun Young Go, Eunhye Jung, Young Eui Jeong, Udeni B. R. Balasuriya
Genome Announcements.2015;[Epub] CrossRef - Comparison of Four Serological Tests for Detecting Antibodies to Japanese Encephalitis Virus after Vaccination in Children
Go Woon Cha, Jung Eun Cho, Young Ran Ju, Young-Jin Hong, Myung Guk Han, Won-Ja Lee, Eui Yul Choi, Young Eui Jeong
Osong Public Health and Research Perspectives.2014; 5(5): 286. CrossRef - Dengue virus type 1 clade replacement in recurring homotypic outbreaks
Boon-Teong Teoh, Sing-Sin Sam, Kim-Kee Tan, Jefree Johari, Meng-Hooi Shu, Mohammed Bashar Danlami, Juraina Abd-Jamil, NorAziyah MatRahim, Nor Muhammad Mahadi, Sazaly AbuBakar
BMC Evolutionary Biology.2013;[Epub] CrossRef - Travel-Associated Chikungunya Cases in South Korea during 2009–2010
Go Woon Cha, Jung Eun Cho, Eun Ju Lee, Young Ran Ju, Myung Guk Han, Chan Park, Young Eui Jeong
Osong Public Health and Research Perspectives.2013; 4(3): 170. CrossRef - The Road Less Traveled
Chaeshin Chu
Osong Public Health and Research Perspectives.2011; 2(1): 1. CrossRef
- Genotypic persistence of dengue virus in the Philippines



 First
First Prev
Prev


