Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 3(4); 2012 > Article
-
Articles
Enhanced Type III Secretion System Expression of AtypicalShigella flexneri II:(3)4,7(8) - Sahyun Honga, Injun Chaa, Nan-Ok Kima, Seong-Han Kima, Kyung-Tae Junga, Je-Hee Leeb, Dong-Wook Kimb, Mi-Sun Parka, Yeon-Ho Kanga
-
Osong Public Health and Research Perspectives 2012;3(4):222-228.
DOI: https://doi.org/10.1016/j.phrp.2012.10.002
Published online: December 31, 2012
aDivision of Enteric Bacterial Infections, Korea National Institute of Health, Osong, Korea.
bMolecular Bacteriology Laboratory, International Vaccine Institute, Seoul, Korea.
- Corresponding author. E-mail: slowpc@daum.net
• Received: August 28, 2012 • Revised: September 26, 2012 • Accepted: October 2, 2012
Copyright ©2012, Korea Centers for Disease Control and Prevention
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
- 2,357 Views
- 17 Download
Abstract
-
Objectives
- We aimed at evaluating the virulence of atypical Shigella flexneri II:(3)4,7(8) by DNA microarray and invasion assay.
-
Methods
- We used a customized S. flexneri DNA microarray to analyze an atypical S. flexneri II:(3)4,7(8) gene expression profile and compared it with that of the S. flexneri 2b strain.
-
Results
- Approximately one-quarter of the atypical S. flexneri II:(3)4,7(8) strain genes showed significantly altered expression profiles; 344 genes were more than two-fold upregulated, and 442 genes were more than 0.5-fold downregulated. The upregulated genes were divided into the category of 21 clusters of orthologous groups (COGs), and the “not in COGs” category included 170 genes. This category had virulence plasmid genes, including the ipa-mxi-spa genes required for invasion of colorectal epithelium (type III secretion system). Quantitative reverse-transcription polymerase chain reaction results also showed the same pattern in two more atypical S. flexneri II:(3)4,7(8) strains. Atypical S. flexneri II:(3)4,7(8) showed four times increased invasion activity in Caco-2 cells than that of typical strains.
-
Conclusion
- Our results provide the intracellularly regulated genes that may be important for adaptation and growth strategies of this atypical S. flexneri.
- Shigella spp. is the causative agent of bacillary dysentery in humans. It is transmitted via the fecal–oral route and causes disease by invading the colonic epithelium, which results in tissue destruction and massive inflammation [1].
- The annual incidence of shigellosis had been estimated to be about 10 cases before 1997, which exploded to about 1000–2500 cases during 1998–2000 in Korea. Furthermore, the annual incidence of shigellosis is increasing gradually in Korea [2]. Three types of serotypically atypical Shigella flexneri strains were isolated in 2007 in our previous study. Among these, the major atypical strain displayed a plural agglutination pattern by reacting with one typing sera (II) and bound with two grouping sera (3)4 and 7(8) (Supplementary Table 1). This atypical strain, II:(3)4, 7(8), showed higher antibiotic resistance to ampicillin, streptomycin, and trimethoprim–sulfamethoxazolethan that of typical S. flexneri, and its resistance is increasing [3,4].
- All pathogenic Shigella strains possess a virulence plasmid that encodes the invasion plasmid antigens IpaACDB and the Mxi-Spa-type III secretion system (TTSS) required for invasion of colorectal epithelium [5,6]. The type III secretion pathway is present in numerous Gram-negative pathogenic bacteria to deliver virulence proteins to the host cell membrane [7,8]. The type III secretion system consists of the following: (1) a secretion apparatus that spans the bacterial envelope; membrane expression of ipa (Mxi) MxiG to MxiM; surface presentation of ipa (spa) Spa13, 24, 29, 32, 33, 40, etc.; (b) translocators and effectors that are inserted into the host cell membrane and act as effectors manipulating host cell processes in favor of the bacteria; invasion plasmid antigens (Ipa) IpaA to IpaD, IpgB1, and IpgD [9,10]; (c) chaperones that associate with the translocators ipgA, ipgC, ipgE, and spa15; and (d) transcriptional activators required for the expression of components of the type III secretion apparatus: VirB and MxiE [11-14].
- Regulation of type III secretion is tightly controlled by the regulators virF and the MxiE regulon, and the pCP301 maintenance system as well as two component systems [15,16]. This type III secretion system is only weakly active during bacterial growth in broth and is activated upon contact of the bacteria with host cells [17].
- Atypical S. flexneri II((3)4,7(8)) also has the Ipa and Mxi-Spa-type III secretion system, but the expression intensity and regulation are still unclear. Consequently, the major aim of this study was to investigate the gene expression profile of atypical S. flexneri.
- It would be useful to characterize the isolated atypical S. flexneri in relation to infections. Additionally, these strains were isolated from children and adults with severe dysentery, emphasizing the need to study these isolates in detail.
1. Introduction
- 2.1. Sample collection
- The tested atypical S. flexneri II:(3)4,7(8) strain was collected from the Korea National Institute of Health. The S. flexneri 2b (ATCC12022) strain was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Serotyping of the S. flexneri strains was confirmed using a commercially available antisera kit (Denka Seiken, Tokyo, Japan) specific for all type- and group-factor antigens. Strains were subcultured on tryptic soy agar (TSA) (Difco, Becton Dickinson and Company, Sparks, MD, USA) plates, and serological reactions were performed after about 18 hours of incubation using the slide agglutination test, according to the manufacturer’s instructions.
- 2.2. DNA microarray and bacterial RNA isolation
- We used a customized S. flexneri DNA microarray from eBiogene (Seoul, Korea). This S. flexneri microarray allowed us to study the expression of 4670 S. flexneri open reading frames. Bacterial RNA was extracted from atypical S. flexneri II:(3)4,7(8), and RNA of the S. flexneri 2b strain grown on TSA for 18 hours was extracted using a Qiagen total RNA purification kit (Qiagen, Valencia, CA, USA). Bacterial RNA was further purified by DNase I treatment and phenol–chloroform extraction. The size of the RNA was confirmed using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
- 2.3. cDNA microarray
- The integrity of bacterial total RNA was checked by capillary electrophoresis with an Agilent 2100 bioanalyzer and further purified with an RNeasy mini kit (Qiagen). cDNA probes for the cDNA microarray analysis were prepared by reverse transcription of total RNA (25 μg) in the presence of aminoallyl-dUTP and 6 μg of random primers (Invitrogen, Carlsbad, CA, USA) for 3 hours. The cDNA probes were cleaned up using a Microcon YM-30 column (Millipore, Bedford, MA, USA), followed by coupling of Cy3 dye (for reference) or Cy5 dye (for test sample) (Amersham Pharmacia, Uppsala, Sweden). The Cy3- or Cy5-labeled cDNA probes were purified with a QIAquick polymerase chain reaction (PCR) purification kit (Qiagen). Dried Cy3- or Cy5-labeled cDNA probes were resuspended in a hybridization buffer containing 30% formamide, 5× SSC, 0.1% SDS, and 0.1 mg/mL salmon sperm DNA. The Cy3- or Cy5-labeled cDNA probes were mixed and hybridized to a microarray slide (S. flexneri microarray) (MYcroarray, Ann Arbor, MI, USA). The slide was washed after an overnight incubation at 42 ℃ and then dried by centrifugation at 650 rpm for 5 minutes. The hybridization image on the slide was scanned with an Axon 4000B instrument (Axon Instruments, Union City, CA, USA).
- 2.4. Data acquisition and microarray data analysis
- After hybridization, the slide was washed and scanned using a GenePix 4000A scanner (Axon Instruments). Fluorescent spots and local background intensities were quantified using Genepix Pro 3.0 software (Axon Instruments) to obtain gene expression ratios (reference vs. test sample). Global lowness was used for data analysis with normalization. The benchmarks for up- and downregulated genes in hybridizations were two- and 0.5-fold, respectively. Gene clustering by gene function was performed at the National Center for Biotechnology Information website by clusters of orthologous groups (COGs) of proteins, http://www.ncbi.nlm.nih.gov/COG [18].
- 2.5. Quantitative real-time reversetranscription polymerase chain reaction
- Microarray data were confirmed by quantitative realtime reverse-transcription polymerase chain reaction (qRT-PCR) using a LightCycler (Roche, Laval, QC, Canada) with software version 5.32 and a Light-Cycler FastStart DNA MasterPlus Syber Green I Master Mix kit (Roche). Total RNA (10 ng) was converted to cDNA using a Qiagen cDNA synthesis kit (Qiagen), following the manufacturer’s instructions. The cDNA was then used as a template for PCR (Taq DNA polymerase, Qiagen). The primers used in the RT-PCR assays are listed in Supplementary Table 2.
- Crossing points were estimated using the “Fit Points Method” software option. Estimates of fold induction were obtained from triplicate samples using RelQuant software (Roche).
- 2.6. Bacterial infection of cultured cells
- Human HeLa cervical epithelial and human Caco-2 colonic epithelial cell lines were grown in Dulbecco’s minimal essential medium. HeLa and Caco-2 cells were seeded at 4 × 105 cells in six-well plates containing 10% fetal bovine serum. They were infected with three atypical and three typical 2b strains, including ATCC12022 at a multiplicity of infection of ~100 per cell, and the plates were centrifuged at 600× g for 10 minutes to synchronize the infection stage. After a 1- hour incubation, gentamicin (50 μg/mL) was added to kill extracellular bacteria, and the cells were then incubated for 1 hour. The cells were washed five times with phosphate buffered saline (PBS) to eliminate viable extracellular bacteria, and trypsinized and lysed in a solution of 0.5% sodium deoxycholate in distilled water. Dilutions of this suspension were then plated onto brain heart infusion agar, and colonies were counted after overnight growth. The number of bacteria per infected cell was counted and averaged. Experiments were repeated at least three times for each strain tested.
2. Materials and Methods
- 3.1. Global transcription profiles
- Comparison of the expression profiles of atypical S. flexneri II:(3)4,7(8) was performed with the typical 2b transcriptome as a control. In total, 4670 expressed genes were categorized by COG of the proteins. We specifically analyzed more than two-fold changed genes. A total of 344 S. flexneri II:(3)4,7(8) genes were more than two-fold upregulated, and 442 genes were 0.5-fold downregulated. These up- and downregulated genes were distributed in 12 COG functional categories (Figure 1A and B). A major category of the upregulated genes was “not in COGs”, which mainly contained ipa-mxi-spa virulence plasmid genes. The second category was “replication and recombination”. The remaining 10 categories showed minor fold changes. Of the downregulated genes, a major category was also “not in COGs”, but it contained only a few virulence genes. The remaining 11 COG distribution categories showed no specific differences.
- 3.2. Expression of virulence plasmid coding genes
- We focused on the “not in COGs” (including virulence plasmid genes) category of the 344 upregulated atypical S. flexneri II:(3)4,7(8) genes, specifically the 41 virulence plasmid coded genes. Most of the genes were invasion-related genes, such as ipa-mxi-spa. Therefore, we specifically surveyed the major regulatory factors of the virulence genes, such as virF, virB, MxiE, the twocomponent signal transduction system, and the global transcription regulators [13] (Table 1).
- 3.3. Expression of virF invasion genes
- The expression of ipa-mxi-spa genes was drastically upregulated in atypical S. flexneri II:(3)4,7(8). The ipa-mxi-spa expression involves a complex regulatory mechanism [19]. Two plasmid-bornegenes, virF and virB, encode essential regulatory proteins. The VirF protein, an AraC-like transcription factor, activates virB and icsA/virG, and the VirB protein, in turn, binds to the promoters of entry genes and activates them [20]. virF and virB expression was observed, and virF and virB showed 52- and 96-fold increases, respectively, compared to that of the ATCC 2b strain.
- 3.4. Expression of the MxiE regulon
- MxiE is a member of the AraC family of regulators that controls a set of late gene products secreted through the Mxi-Spa TTSS [13]. The MxiE regulon is upregulated after entry into epithelial cells and includes virA, ipaH7.8 (involved in the escape of Shigella into the host cytosol), and ipaH9.8 (the product of which is targeted to the host nucleus) [21]. MxiE showed the highest expression (140-fold increase), and virA expression was 14.1-fold upregulated.
- 3.5. Maintenance of virulence plasmid pCP301
- The R100 plasmid is the primary replication system of the virulence plasmid, which belongs to the RepFIIA family of replicons [16]. There are also multiple loci on pCP301 that are homologous to sequences known to be involved in plasmid segregation and stable maintenance such as stbA and stbB of plasmid R100 and mvpA and mvpT. Among these, mvpA and mvpT form an effective system to maintain stability in vitro [15]. The expression of mvpA was not significantly altered (1.2-fold change), whereas mvpT increased 26-fold compared to that in the control strain.
- Gene products influencing expression of Shigella flexneri plasmid-linked virulence genes
- 3.6. Two-component signal transduction systems, sigma factors, and stress response factors
- Two-component signal transduction systems play an important role in regulating virulence in S. flexneri. CpxR binds to the virF promoter region when it is phosphorylated by CpxA [22]. Expressions of cpxA (1.4- fold increase) and cpxB (0.5-fold decrease) were not significantly changed.
- The well-characterized PhoP–PhoQ system is crucial for persistent infection and resistance to killing by cationic polypeptides derived from polymorphonuclear leukocytes and other sources [23]. We observed the transcription of these and other two-component systems; PhoP changed 0.8-fold and PhoQ 0.6-fold. Changes were observed for the ompR–envZ osmosensor system, as ompR changed 0.2-fold and envZ 1.3-fold.
- Expressions of the rpoS gene, the master regulator of general stress, and sigma factor were not altered significantly.
- H-NS, global regulatory factor hns represses the virB gene antagonistically. A footprint extending from position –20 to position +20 has been detected in vitro at the virB promoter, suggesting that H-NS may exclude RNA polymerase from binding there [24,25]. The hns changed 1.16-fold and its analog, stpA, changed 1.89- fold.
- 3.7. Other regulators
- It appeared that the expression of the virulence genes may be coupled to the bacterial cell cycle, as ispA, a cell division gene, changed 0.68-fold [26].
- The mia gene, which codes for the tRNAN6-isopenthyladenosine (i6A37) synthetase, is required for production of the modified nucleoside 2-methyl-N6- isopentyladenosine necessary for expressing the virulence regulon [27]. This gene changed 0.74-fold.
- It seemed that the alteration in DNA topology facilitated a productive interaction between the DNA-bound VirF protein and RNA polymerase due to changes in pH, temperature, and osmolarity. Thus, the change in DNA supercoiling may promote VirF oligomerization on the DNA, and this may represent an activation step analogous to arabinose binding to the AraC protein. Thus, topA (0.56-fold change) [28], the gene encoding DNA topoisomerase I, and parC (0.95) and ParE (1.16), the genes encoding DNA topoisomerase IV, facilitate virulence gene expression [29]. All these regulators changed <2-fold.
- 3.8. Validation of the microarray results by qRT-PCR
- We confirmed the array experiment by qRT-PCR. Regulatory genes of type III secretion systems were selected and compared. RNA samples were isolated under the same conditions as the microarray experiment, and the qRT-PCR results were compared with the microarray data (Table 1). The qRT-PCR results showed a similar pattern as that of the array experiment. We also tested two more atypical strains to confirm high expression of the type III secretion system. The results showed the same pattern. The primers used are listed in Supplementary Table 2.
- 3.9. Bacterial infection of cultured cells
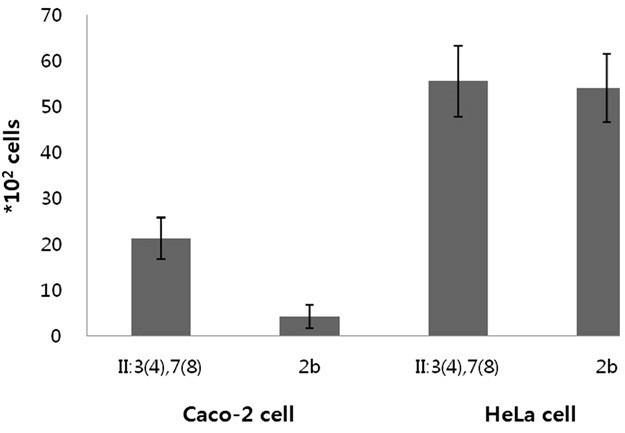
- We questioned the invasion ability of these atypical strains based on the increased expression of the type III secretion system. Invasion of three atypical S. flexneri II:(3)4,7(8) and three typical 2b (including ATCC 12022) were estimated in colon carcinoma Caco-2 cells and epithelial HeLa cells. The atypical S. flexneri II:(3) 4,7(8) strains demonstrated more than four-fold increased ability of invasion compared to that of typical 2b strains, specifically in Caco-2 cells but not in HeLa cells (Figure 2).
3. Results
Figure 1.
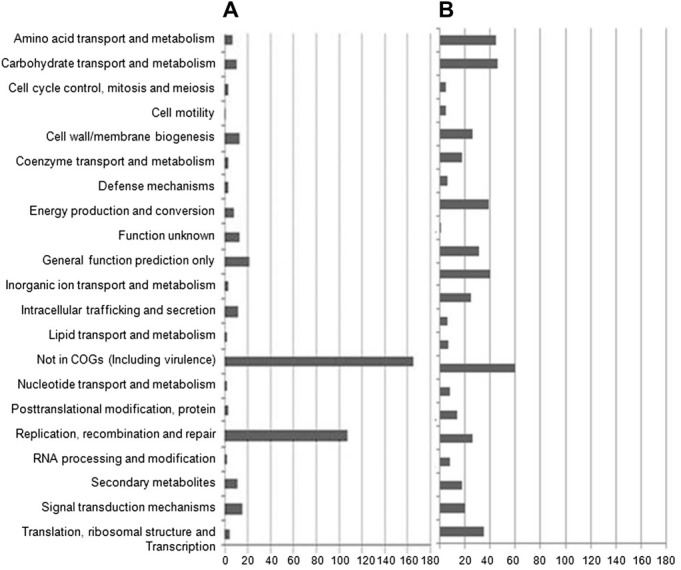
Global transcription profiles of an atypical Shigella flexneri strain. (A) More than two-fold upregulated genes were distributed as COGs. (B) More than two-fold downregulated genes were distributed as COGs. X axis indicates the number of genes. COGs = clusters of orthologous groups.

Table 1.
- In this study, we analyzed the gene expression profile of atypical S. flexneri II:(3)4,7(8) and compared it to that of ATCC 2b as a control strain. We analyzed more than two-fold changed genes. Of the two-fold upregulated genes, the major category of 344 genes was “not in COGs”. The “not in COGs” (including virulence plasmid genes) category contained 41 invasion-related genes of the ipa-mxi-spatype III secretion system. Among the regulatory genes for the type III secretion system, virF and the MxiE regulon were specifically upregulated in atypical S. flexneri II:(3)4,7(8). We also confirmed expression of these regulator genes by qRT-PCR in two more atypical S. flexneri II:(3)4,7(8) and two more 2b serotype strains. Additionally, atypical S. flexneri II:(3)4,7(8) showed a four-fold increased invasion activity in Caco-2 cells compared to that of typical strains but not in HeLa cells. In the human colonic epithelial cell line Caco-2, which differentiates into a polarized epithelium expressing a well-established brush border, the invasion process occurs through basolateral surfaces and has been used in an invasion study [30,31].
- The type III secretion system is regulated at several levels. First, transcription of secretion genes is regulated by global regulators, such as H-NS, IspA, Mia, ParC, and ParE, which respond to changes in temperature, pH, or osmolarity [32]. These global regulators did not show specific changes in the atypical S. flexneri II:(3)4,7(8) strains. Thus, increased expression of the type III secretion system in atypical strains was low in relation to global regulation. Next, activity of the type III secretion system apparatus is regulated in response to specific stimuli, including contact with host cells, exposure to artificial compounds such as Congo red, and alterations in the growth environment. These stimuli were not altered in atypical S. flexneri II:(3)4,7(8) strains or in the control strain. Thus, various stimuli did not increase expression of the type III secretion system in the atypical strains. Finally, transcriptional activators such as VirB and MxiE were required for proper expression of the type III secretion apparatus. The atypical S. flexneri II:(3)4,7(8) strains showed more than 100-fold increases in MxiE and a 96-fold increase in VirB expression compared to those of the control strain; thus, increased type III secretion system expression of the atypical strain originated from increased MxiE and VirB transcription. Most Shigella spp. show low transcription levels of these regulators and increase only when they contact host epithelial cells or are exposed to artificial compounds such as Congo red. Other reports have shown that increased secretion occurs when some genes such as ipaB and ipaD are inactivated, resulting in deregulated secretion [17,33]. Inducing or deregulating secretion activity also results in increased transcription of some genes encoding secreted proteins, such as the virA and ipaH genes in Shigella spp. [13]. In atypical S. flexneri II:(3)4,7(8), the type III secretion system was induced by an increased expression of the MxiE regulon, but the exact reason for the increased expression of the virulence-related genes is still uncertain in atypical S. flexneri II:(3)4,7(8). Increased type III secretion system expression and increased invasion ability of human epithelial cells of these atypical S. flexneri II:(3)4,7(8) strains will be studied specifically in the near future, because this atypical S. flexneri II:(3)4,7(8) strain is increasing in prevalence in Korea and shows increased antibiotic resistance.
- These results of this study will facilitate functional studies of intracellularly regulated genes that may be important for adaptation and growth strategies of this atypical S. flexneri during infection.
4. Discussion
- Supplementary data associated with this article can be found in online version at http://dx.doi.org/10.1016/j.phrp.2012.10.002.
Appendix A. Supplementary material
-
Acknowledgements
- This study was supported by an intramural grant from the Korean National Institute of Health, Republic of Korea (Grant: 4851-300-210-13).
- 1. Labrec EH Schneider H Magnani TJ et al.. Epithelial cell penetration as an essential step in the pathogenesis of bacillary dysentery. J Bacteriol 11;1964;88(5). 1503−18. PMID: 16562000.ArticlePubMedPMC
- 2. Cho SH Kim JH Kim JC et al.. Surveillance of bacterial pathogens associated with acute diarrheal disease in the Republic of Korea during one year. J Microbiol 6;2003;44(3). 327−35. PMID: 16820763.
- 3. Hong S Choi YH Choo YA et al.. Genetic characterization of atypical Shigella flexneri isolated in Korea. J Microbiol Biotechnol 10;2010;20(10). 1457−62. PMID: 21030833.ArticlePubMed
- 4. Dorman CJ Porter ME . The Shigella virulence gene regulatory cascade: a paradigm of bacterial gene control mechanisms. Mol Microbiol 8;1998;29(3). 677−84. PMID: 9723908.ArticlePubMed
- 5. Andrews GP Hromockyj AE Coker C et al.. Two novel virulence loci, mxiA and mxiB, in Shigella flexneri 2a facilitate excretion of invasion plasmid antigens. Infect Immun 6;1991;59(6). 1997−2005. PMID: 2037361.ArticlePubMedPMC
- 6. Sansonetti PJ Kopecko DJ Formal SB . Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun 3;1982;35(3). 852−60. PMID: 6279518.ArticlePubMedPMC
- 7. Lucchini S Liu H Jin Q et al.. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect Immun 1;2005;73(1). 88−102. PMID: 15618144.ArticlePubMedPMC
- 8. Talukder KA Islam MA Dutta DK et al.. Phenotypic and genotypic characterization of serologically atypical strains of Shigella flexneri type 4 isolated in Dhaka, Bangladesh. J Clin Microbiol 7;2002;40(7). 2490−7. PMID: 12089268.ArticlePubMedPMC
- 9. Skoudy A Mounier J Aruffo A et al.. CD44 binds to the Shigella IpaB protein and participates in bacterial invasion of epithelial cells. Cell Microbiol 2;2000;2(1). 19−33. PMID: 11207560.ArticlePubMed
- 10. High N Mounier J Prevost MC et al.. IpaB of Shigella flexneri causes entry into epithelial cells and escape from the phagocytic vacuole. EMBO J 1992;11(5). 1991−9. PMID: 1582426.ArticlePubMedPMC
- 11. Hueck CJ . Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 6;1998;62(2). 379−433. PMID: 9618447.ArticlePubMedPMC
- 12. Mavris M Sansonetti PJ Parsot C . Identification of the cis-acting site involved in activation of promoters regulated by activity of the type III secretion apparatus in Shigella flexneri. J Bacteriol 12;2002;184(24). 6751−9. PMID: 12446624.ArticlePubMedPMC
- 13. Demers B Sansonetti PJ Parsot C . Induction of type III secretion in Shigella flexneri is associated with differential control of transcription of genes encoding secreted proteins. EMBO J 5;1998;17(10). 2894−903. PMID: 9582283.ArticlePubMedPMC
- 14. Kuwae A Yoshida S Tamano K et al. Shigella invasion of macrophage requires the insertion of IpaC into the host plasma membrane. Functional analysis of IpaC. J Biol Chem 8;2001;276(34). 32230−9. PMID: 11413141.ArticlePubMed
- 15. Sayeed S Reaves L Radnedge L et al.. The stability region of the large virulence plasmid of Shigella flexneri encodes an efficient postsegregational killing system. J Bacteriol 5;2000;182(9). 2416−21. PMID: 10762240.ArticlePubMedPMC
- 16. Venkatesan MM Goldberg MB Rose DJ et al.. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect Immun 5;2001;69(5). 3271−85. PMID: 11292750.ArticlePubMedPMC
- 17. Menard R Sansonetti P Parsot C . The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J 11;1994;13(22). 5293−302. PMID: 7957095.ArticlePubMedPMC
- 18. Tatusov RL Galperin MY Natale DA et al.. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 1;2000;28(1). 33−6. PMID: 10592175.ArticlePubMedPMC
- 19. Dorman CJ McKenna S Beloin C . Regulation of virulence gene expression in Shigella flexneri, a facultative intracellular pathogen. Regulation of virulence gene 5;2001;291(2). 89−96.Article
- 20. Sandlin RC Lampel KA Keasler SP et al.. Avirulence of rough mutants of Shigella flexneri: requirement of O antigen for correct unipolar localization of IcsA in the bacterial outer membrane. Infect Immun 1;1995;63(1). 229−37. PMID: 7528731.ArticlePubMedPMC
- 21. Kane CD Schuch R Day Jr WA et al.. MxiE regulates intracellular expression of factors secreted by the Shigella flexneri 2a type III secretion system. J Bacteriol 8;2002;184(16). 4409−19. PMID: 12142411.ArticlePubMedPMC
- 22. Nakayama S Watanabe H . Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J Bacteriol 7;1998;180(14). 3522−8. PMID: 9657992.ArticlePubMedPMC
- 23. Moss JE Fisher PE Vick B et al.. The regulatory protein PhoP controls susceptibility to the host inflammatory response in Shigella flexneri. Cell Microbiol 12;2000;2(6). 443−52. PMID: 11207599.ArticlePubMed
- 24. Dorman CJ Ni Bhriain N Higgins CF . DNA supercoiling and environmental regulation of virulence gene expression in Shigella flexneri. Nature 4;1990;344(6268). 789−92. PMID: 2184366.ArticlePubMed
- 25. Tobe T Yoshikawa M Mizuno T et al.. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J Bacteriol 10;1993;175(19). 6142−9. PMID: 7691791.ArticlePubMedPMC
- 26. Síomóin RA Nakata N Murai T et al.. Identification and characterization of ispA, a Shigella flexneri chromosomal gene essential for normal in vivo cell division and intercellular spreading. Mol Microbiol 2;1996;19(3). 599−609. PMID: 8830250.ArticlePubMed
- 27. Durand JM Bjork GR Kuwae A et al.. The modified nucleoside 2- methylthio-N6-isopentenyladenosine in tRNA of Shigella flexneri is required for expression of virulence genes. J Bacteriol 9;1997;179(18). 5777−82. PMID: 9294434.ArticlePubMedPMC
- 28. Ni Bhriain N Dorman CJ . Isolation and characterization of a topA mutant of Shigella flexneri. Mol Microbiol 2;1993;7(3). 351−8. PMID: 8384681.ArticlePubMed
- 29. McNairn E Ni Bhriain N Dorman CJ . Overexpression of the Shigella flexneri genes coding for DNA topoisomerase IV compensates for loss of DNA topoisomerase I: effect on virulence gene expression. Mol Microbiol 2;1995;15(3). 507−17. PMID: 7783621.ArticlePubMed
- 30. Mounier J Vasselon T Hellio R et al.. Shigella flexneri enters human colonic Caco-2 epithelial cells through the basolateral pole. Infect Immun 1;1992;60(1). 237−48. PMID: 1729185.ArticlePubMedPMC
- 31. Talukder KA Islam MA Khajanchi BK et al.. Temporal shifts in the dominance of serotypes of Shigella dysenteriae from 1999 to 2002 in Dhaka, Bangladesh. J Clin Microbiol. 11;2003;41(11). 5053−8. PMID: 14605138.ArticlePubMedPMC
- 32. Friedberg D Umanski T Fang Y et al.. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol Microbiol 12;1999;34(5). 941−52. PMID: 10594820.ArticlePubMed
- 33. Menard R Sansonetti PJ Parsot C . Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol 9;1993;175(18). 5899−906. PMID: 8376337.ArticlePubMedPMC
Figure & Data
References
Citations
Citations to this article as recorded by 



 PubReader
PubReader Cite
Cite
