Articles
- Page Path
- HOME > Osong Public Health Res Perspect > Volume 7(4); 2016 > Article
-
Original Article
Distribution of Antibodies Specific to the 19-kDa and 33-kDa Fragments ofPlasmodium vivax Merozoite Surface Protein 1 in Two Pathogenic Strains Infecting Korean Vivax Malaria Patients - Sylvatrie-Danne Dinzouna-Boutambaa, Sanghyun Leeb, Ui-Han Sona,c, Su-Min Songa, Hye Soo Yuna, So-Young Jooa, Dongmi Kwakc, Man Hee Rheec, Dong-Il Chunga, Yeonchul Honga, Youn-Kyoung Gooa
-
Osong Public Health and Research Perspectives 2016;7(4):213-219.
DOI: https://doi.org/10.1016/j.phrp.2016.05.006
Published online: June 25, 2016
aDepartment of Parasitology and Tropical Medicine, Kyungpook National University School of Medicine, Daegu, Korea
bDivision of Biosafety Evaluation and Control, Korea National Institute of Health, Cheongju, Korea
cCollege of Veterinary Medicine, Kyungpook National University, Daegu, Korea
- ∗Corresponding author. kuku1819@knu.ac.kr
- 1These authors contributed equally to this work.
• Received: April 28, 2016 • Revised: May 23, 2016 • Accepted: May 30, 2016
Copyright © 2016 Korea Centers for Disease Control and Prevention. Published by Elsevier Korea LLC.
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Abstract
-
Objectives
- Plasmodium vivax merozoite surface protein 1 (PvMSP1) is the most intensively studied malaria vaccine candidate. Although high antibody response-inducing two C-terminal fragments of PvMSP1 (PvMSP1-19 and PvMSP1-42) are currently being developed as candidate malaria vaccine antigens, their high genetic diversity in various isolates is a major hurdle. The sequence polymorphism of PvMSP1 has been investigated; however, the humoral immune responses induced by different portions of this protein have not been evaluated in Korea.
-
Methods
- Two fragments of PvMSP1 were selected for this study: (1) PvMSP1-19, which is genetically conserved; and (2) PvMSP1-33, which corresponds to a variable portion. For the latter, two representative strains, Sal 1 and Belem, were included. Thus, three recombinant proteins, PvMSP1-19, PvMSP1-33 Sal 1, and PvMSP1-33 Belem, were produced in Escherichia coli and then tested by enzyme-linked immunosorbent assays using sera from 221 patients with vivax malaria.
-
Results
- Of the 221 samples, 198, 142, and 106 samples were seropositive for PvMSP1-19, PvMSP1-33 Sal 1, and PvMSP1-33 Belem, respectively. Although 100 samples were simultaneously seropositive for antibodies specific to all the recombinant proteins, 39 and six samples were respectively seropositive for antibodies specific to MSP1-33 Sal 1 and MSP1-33 Belem. Antibodies specific to PvMSP1-19 were the most prevalent.
-
Conclusion
- Monitoring seroprevalence is essential for the selection of promising vaccine candidates as most of the antigenic proteins in P. vivax are highly polymorphic.
- Plasmodium vivax is the most prevalent species that causes malaria in humans [1]. It is endemic in the tropical and subtropical countries of Africa, the Middle East, the South Pacific, Central and South America, and in Asia, including the Republic of Korea (ROK) 2, 3. In recent years, several reports throughout the world have linked P. vivax to severe disease and death 4, 5, 6. These findings associated with the emergence of drug-resistant strains have increased concerns regarding this species [7]. Since an effective malaria vaccine capable of inducing robust and long-lasting protection in naturally exposed individuals would be an important tool for malaria control, studies evaluating immune responses against different P. vivax vaccine candidates are urgently required.
- Proteins expressed on the surface of P. vivax merozoites are important candidates for malaria vaccine development. Among these proteins, merozoite surface protein 1 (MSP1) is the most intensively studied. MSP1 is synthesized as a high molecular weight precursor (approximately 200 kDa), which is then processed into several smaller MSPs [8]. During invasion, the C-terminal 42-kDa fragment (MSP1-42) is further processed into 33-kDa (MSP1-33) and 19-kDa (MSP1-19) fragments. Only the MSP1-19 fragment remains on the merozoite surface and is transported into the invaded erythrocytes 9, 10. The C-terminus of MSP1 reportedly induces high antibody responses in hosts, and specific antibodies against this region are known to inhibit merozoite invasion 11, 12. Although both MSP1-19 and MSP1-42 are being considered as potential vaccine candidates, the processing and presentation of these fragments may be problematic due to the large number of disulfide linkages in the two epidermal growth factor-like regions of MSP1-19 13, 14. In addition, the MSP1-33 fragment, which is the fragment of MSP1-42 without MSP1-19, shows an extensive polymorphism in malaria patient populations [15]. Three representative MSP1 variants of P. vivax (PvMSP1)—Belem, Sal-1, and recombinant types—have been observed in the ROK 16, 17. In addition, single-nucleotide polymorphisms have frequently been observed in P. vivax isolates from vivax malaria patients [15]. Studies on the MSP1 polymorphism have been performed in the ROK; however, the distribution of strain-specific antibodies has not yet been evaluated 18, 19. In this study, we generated three recombinant proteins of which two correspond to the polymorphic variants of PvMSP1-33 (PvMSP1-33 Sal 1 and PvMSP1-33 Belem) and the other corresponds to the conserved PvMSP1-19. We also evaluated antibody responses to these proteins in individuals infected with P. vivax in ROK to determine the frequency and the magnitude of the humoral response against different P. vivax vaccine candidate antigens.
Introduction
- 2.1 Ethics statement
- This study was approved by the research ethics committee of Kyungpook National University (Daegu, Korea). All the participants signed written informed consent forms and agreed to provide 5-mL blood samples.
- 2.2 Sample collection
- The samples were collected at hospitals and health centers throughout the northern region of the ROK, where vivax malaria is endemic in the summer season (June to August). In 2015, 90.4% (619/685) of vivax malaria cases reported in ROK had occurred in this area. Venous blood samples with EDTA were obtained from 221 individuals showing classic symptoms of malaria, who sought treatment at the health facilities mentioned below. The samples were first diagnosed as vivax malaria using a rapid diagnostic test kit (NanoSign Malaria P.f/P.v; Bioland, Seoul, Korea) at a hospital or health center. After blood collection and diagnosis, all the patients were treated with chloroquine. First of all, 600 mg chloroquine was administered, and then three more doses of 300 mg chloroquine were administered at 6 hours, 24 hours, and 48 hours after the first dose.
- The blood samples were centrifuged at 1,500g for 15 minutes to obtain plasma for further studies. The plasma samples were transported on ice to a laboratory in the Department of Parasitology and Tropical Medicine at Kyungpook National University and were stored at −70°C until required. The 221 blood samples from vivax malaria patients consisted of 140 and 81 samples collected in 2012 and 2013, respectively. All the samples were collected from symptomatic vivax malaria patients who had not been previously infected with P. vivax. The mean age of the patients at sampling in 2012 and 2013 was 43.2 years and 36.4 years, respectively. Of all patients from whom samples were collected from in 2012 and 2013, 40.0% (56/140) and 30.9% (25/81) were from men, respectively. In most samples, the P. vivax parasite was not observed using microscopy due to low parasitemia, even though the rapid diagnostic test and nested-polymerase chain reaction (PCR) were positive. Samples were also obtained from 30 healthy volunteers who resided in nonendemic areas (southern parts) of the ROK and had not traveled to vivax malaria endemic areas.
- 2.3 Microscopic examination and nested-PCR for vivax malaria diagnosis
- All the blood samples were examined using microscopic analysis of Giemsa-stained thick and thin blood films and using nested-PCR, targeting the 18S ribosomal RNA as described previously 20, 21. Briefly, 5 μL of extracted DNA was used as the template in a primary PCR, and 1 μL of the primary PCR product was used as a template in the second amplification. The cycling conditions for the primary PCR were as follows: initial denaturation at 94°C for 5 minutes, followed by 25 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 1 minute, and extension at 72°C for 1 minute. The cycling conditions for the second PCR were: initial denaturation at 92°C for 5 minutes, followed by 25 cycles of denaturation at 92°C for 20 seconds, annealing at 52°C for 20 seconds, and extension at 78°C for 20 seconds. The final PCR products were detected by staining with ethidium bromide.
- 2.4 PvMSP1 cloning
- Genomic DNA was extracted from a Korean P. vivax isolate using the QIAamp DNA blood kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. The extracted DNA was used to amplify the MSP1-33 Sal 1, MSP1-33 Belem, and MSP1-19 genes. Oligonucleotide primer sets containing BamHI and EcoRI restriction enzyme sites were designed based on the following reference genes: PvMSP1 Sal 1 strain XM001614792 for PvMSP1-33 Sal 1 and PvMSP1-19, and PvMSP1 Belem strain AF435594 for PvMSP1-33 Belem. The following oligonucleotide primers were used: for PvMSP1-19 (450 bp), forward primer 5′-CGTGGATCCAATGAAGTGAAGTCTTCTG-3′ and reverse primer 5′-TTCGAATTCAAGCTCCATGCACAGGAG-3′; for PvMSP1-33 Sal 1 (854 bp), forward primer 5′-GCGGGATCCGTTAAGGAAGCTTTGCAAG-3′ and reverse primer 5′-CGCGAATTCGCAATAAATTCGTTCTTTTTAC-3′; and for PvMSP1-33 Belem (993 bp), forward primer 5′-CGTGGATCCATAAGCTACCTGTCCAGTG-3′ and reverse primer 5′-CGCGAATTCCATCGATTGGGTCTTTTCAGTAG-3’. Amplification was performed in 50 μL 1 × PCR buffer (Takara, Tokyo, Japan) containing 2.5 U Taq polymerase, 2 μL DNA template, 10 pmol of each primer, and 2mM of each deoxynucleoside triphosphate. The cycling conditions were as follows: initial denaturation at 95°C for 5 minutes, followed by 35 cycles of 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds, and a final extension step at 72°C for 5 minutes. The PCR product was cloned into the BamHI and EcoRI restriction enzyme sites of the pGEM-4T-1 vector. The resulting plasmid construct was confirmed with restriction enzyme digestion and nucleotide sequencing. PvMSP1-33 Sal 1 and PvMSP1-33 Belem constructs were selected based on sequencing and Basic Local Alignment Search Tool search results.
- 2.5 Recombinant PvMSP1 expression and purification
- PvMSP1-33 Sal 1, PvMSP1-33 Belem, and PvMSP1-19 were expressed individually as glutathione S-transferase (GST) fusion proteins in the Escherichia coli BL21 (DE3) strain (GE Healthcare, Chicago, IL, USA) according to the manufacturer's instructions. E. coli cells expressing these proteins were washed three times with phosphate-buffered saline (PBS), lysed in 1% Triton X-100-PBS, sonicated, and then centrifuged at 10,000g for 10 minutes at 4°C. Soluble GST-fused rPvMSP1-33 Sal1, rPvMSP1-33 Belem, or rPvMSP1-19 proteins were purified from the cell lysates with glutathione-Sepharose 4B beads (GE Healthcare) according to the manufacturer's instructions. After purification, the GST tag was subsequently removed from the GST-fusion proteins using a Thrombin Kit (Novagen, Darmstadt, Germany).
- 2.6 Sodium dodecyl sulfate polyacrylamide gel electrophoresis and Western blotting
- The expression of recombinant proteins was verified using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining, and the antigenicity of the expressed recombinant proteins was confirmed using Western blot analysis. To identify the antigenicity of the recombinant proteins, we applied pooled serum from 30 patients infected with P. vivax to the Western blot. In addition, 10 patients each infected with P. vivax classified as MSP1 Sal 1 and MSP1 Belem in a previous study [18] were used for another Western blot analysis in order to determine whether the two antigens, PvMSP1-32 Sal 1 and Belem, could distinguish the respective sera. The recombinant proteins were separated by 14% SDS-PAGE and then electroblotted onto a nitrocellulose membrane [22]. The membranes were blocked with 5% skimmed milk in PBS plus 0.05% Tween 20 and then probed with pooled serum (1:100) as the primary antibody. After the membrane was washed with PBS plus Tween 20, horseradish peroxidase-conjugated antihuman immunoglobulin G (1:2,000; Bethyl Laboratories, Montgomery, TX, USA) was applied as the secondary antibody. Finally, the reacted proteins were detected via chemiluminescence using enhanced chemiluminescence Western blotting detection reagents (Amersham Biosciences, Piscataway, NJ, USA).
- 2.7 Enzyme-linked immunosorbent assay
- The optimal concentrations or dilutions of the antigen, serum, enzyme–antibody conjugate, and substrate solutions were determined through “checkerboard” titrations of each reagent against all other reagents. Individual wells of a microtiter plate (Nunc, Roskilde, Denmark) were coated overnight at 4°C with the purified rPvMSP1-19, rPvMSP1-33 Sal1, and rPvMSP1-33 Belem proteins (1 mg/mL) dissolved in 0.05M carbonate-bicarbonate buffer (pH 9.6). The plates were then blocked with 3% skim milk solution for 1 hour at 37°C. After washing with 0.05% Tween 20 in PBS, the plates were incubated with sera at a dilution of 1:100. The binding of the antibody was detected via incubation with horseradish peroxidase-conjugated antihuman immunoglobulin G (IgG, 1:5,000; Bethyl Laboratories) or antihuman IgG1, IgG2, IgG3 (1:5,000; MyBioSource, San Diego, CA, USA) and 2,20-asinobis (3-ethylbenzthiazolne sulfonic acid; Sigma-Aldrich, St. Louis, MO, USA). The optical density (OD) was measured at 415 nm using an MTP-500 microplate reader (Corona Electric, Ibaraki, Japan). The mean OD values of triplicate enzyme-linked immunosorbent assay (ELISA) results were then calculated. The ELISA titer was considered as positive when the OD was ≥ 0.08, ≥ 0.1, and ≥ 0.17 for rPvMSP1-19, rPvMSP1-33 Sal 1, and rPvMSP1-33 Belem, respectively. The OD cutoff values (0.08, 0.1, and 0.17) were calculated by determining the mean OD for normal sera (sera from 30 healthy people) plus threefold standard deviation.
- 2.8 Genetic polymorphisms of the PvMSP1 gene
- The PvMSP1 sequences in the study samples were obtained from a previous study wherein the same samples were classified by the PvMSP1 genotype as PvMSP1 Sal 1, Belem, or recombinant type [18].
- 2.9 Statistical analysis
- The means and standard deviations were calculated, and the statistical significance of the differences was determined by Student t test using JMP Version 8 (SAS Institute Inc., Cary, NC, USA). Differences in the means were considered as statistically significant when the p values were < 0.05.
Materials and methods
- 3.1 Production of PvMSP1-19, PvMSP1-33 Sal 1, and PvMSP1-33 Belem
- Three PvMSP1 proteins, PvMSP1-19, PvMSP1-33 Sal 1, and PvMSP1-33 Belem, were selected for this study (Figure 1A), with the lengths of the nucleotide sequences of the respective coding regions being 450 bp, 854 bp, and 993 bp (data not shown). PvMSP1-19, PvMSP1-33 Sal 1, and PvMSP1-33 Belem were expressed as soluble GST fusion proteins in E. coli, with molecular masses of approximately 42 kDa, 57 kDa, and 61 kDa, respectively. Subsequently, 16-kDa, 31-kDa, and 35-kDa proteins of PvMSP1-19, PvMSP1-33 Sal 1, and PvMSP1-33 Belem, respectively, were produced upon GST cleavage by thrombin (Figure 1B).
- 3.2 Reactivity of PvMSP1-19, PvMSP1-33 Sal 1, and PvMSP1-33 Belem
- To evaluate the antigenicity of PvMSP1-19, PvMSP1-33 Sal 1, and PvMSP1-33 Belem, we performed Western blotting with the three recombinant proteins and sera from patients with vivax malaria and healthy individuals. The recombinant proteins reacted with the sera from the infected patients but not with those from healthy individuals, indicating that the recombinant proteins could be used to detect P. vivax infection in a serum sample (Figure 1C).
- Subsequently, another Western blot analysis was performed to confirm whether rPvMSP1-33 Sal 1 and rPvMSP1-33 Belem could differentiate the sera from patients infected with P. vivax Sal 1 and Belem strains. Infection with P. vivax Sal 1 and Belem was determined based on data from our previous study where infection with these strains was detected by sequencing the PvMSP1 gene [18]. The results showed that the serum from patients infected with P. vivax MSP Sal 1 reacted to rPvMSP1-33 Sal 1 but not to rPvMSP1-33 Belem. In contrast, rPvMSP1-33 Belem reacted to the serum collected from a patient infected P. vivax Belem, but not to rPvMSP1-33 Sal 1 (Figure 1C).
- 3.3 Prevalence of antibodies specific to PvMSP1-19, PvMSP1-33 Sal 1, and PvMSP1-33 Belem
- Among the 221 samples from patients with confirmed vivax malaria, 198 (89.6%), 142 (64.3%), and 106 (48.0%) samples were seropositive for PvMSP1-19, PvMSP1-33 Sal 1, and PvMSP1-33 Belem, respectively (Table 1). In addition, the levels of PvMSP1-19 antibodies were significantly higher than the levels of PvMSP1-33 Sal 1 and Belem antibodies (Figure 2). However, the percentage of samples that were seronegative for PvMSP1-33 Sal 1 (35.7%) and PvMSP1-33 Belem (52.0%) was higher than that for PvMSP1-19 (10.4%). Moreover, 100 (45.3%) samples were seropositive for antibodies to all three recombinant proteins (Table 2).
Results
- Surveillance studies for the prevalence of antibodies specific to vaccine candidates must be performed before selecting a candidate for the development of serodiagnostic tools and vaccines, especially since most of the promising vaccine and serodiagnostic candidates are highly polymorphic. Therefore, this study investigated the prevalence of antibodies specific to PvMSP1-19, PvMSP1-33 Sal 1, and PvMSP1-33 Belem, which are promising vaccine candidates, in Korean P. vivax isolates.
- In Western blotting analyses, the three recombinant proteins successfully demonstrated antigenicity. The antibody response to a Plasmodium antigen depends on the correct folding of conformational epitopes in the antigen. Therefore, in order to investigate the prevalence of antibodies to PvMSP1-19, PvMSP1-33 Sal 1, and PvMSP1-33 Belem, the recombinant proteins should be structurally similar to the native proteins. Previously, recombinant PvMSP1 fragments were used to determine the tertiary structure of native PvMSP1 [23]. Moreover, several studies on the prevalence of specific antibodies to these PvMSP1 fragments have been conducted 12, 24, 25. Therefore, the recombinant proteins generated in this study, were used to evaluate the antibody prevalence in patients with vivax malaria from the ROK.
- Our ELISA results showed that the levels of PvMSP1-19 antibodies were significantly higher than the levels of PvMSP1-33 Sal 1 and Belem antibodies. This may be caused by the presence of two cysteine-rich and immunogenic epidermal growth factor-like domains in PvMSP1-19. Nonetheless, more samples were seronegative for PvMSP1-33 Sal 1 and PvMSP1-33 Belem compared with that for PvMSP1-19, possibly due to a hypervariable region in the PvMSP1-33 fragments. In addition, approximately 100 patients possessed more than two types of PvMSP1-specific antibodies to different strains of PvMSP1, namely PvMSP1 Sal 1 and Belem. These results are consistent with previous studies that showed high multiplicities of infection for P. vivax in Korea 26, 27. However, these studies did not evaluate whether the high multiplicity of infection, as determined with nucleotide sequencing, results in the production of antibodies specific to each strain. In addition, 39 (17.6%) and six (2.7%) PvMSP1-19-seropositive serum samples were also seropositive for PvMSP1-33 Sal 1 and PvMSP1-33 Belem, respectively (Table 2). In a previous study by Kang et al [15], Korean P. vivax isolates showed high genetic diversity in the PvMSP1-33 region. In addition, PvMSP1 has been classified not only as Sal 1 and Belem, but also as recombinant and mutant type [28]. This high genetic diversity might result in seropositivity for PvMSP1-19 and seronegativity for PvMSP1-33 Sal 1 and PvMSP1-33 Belem.
- PvMSP1-19 is a C-terminal fragment located in a conserved region of PvMSP1. Current results showed that PvMSP1-19 is highly antigenic, and that antibodies specific to PvMSP1-19 are the most prevalent antibodies in Korean patients with P. vivax. In addition, seroprevalence studies for specific antibodies to the candidates of the vaccine and serodiagnostic tools are crucial for the development of these preventive and diagnostic strategies.
Discussion
- The authors declare that there are no conflicts of interest regarding the publication of this paper.
Conflicts of interest
-
Acknowledgements
- This research was supported by the Kyungpook National University Research Fund, 2013.
Acknowledgments
- 1. http://apps.who.int/iris/bitstream/10665/97008/1/9789241564694_eng.pdf.
- 2. Battle K.E., Guerra C.A., Golding N.. Global database of matched Plasmodium falciparum and P. vivax incidence and prevalence records from 1985 to 2013. Sci Data 2:2015 Aug 18;150012eCollection 2015. PMID: 26306203.Article
- 3. Gething P.W., Elyazar I.R., Moyes C.L.. A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl Trop Dis 6(9). 2012 Sep;e1814PMID: 22970336.ArticlePubMed
- 4. Baird J.K.. Evidence and implications of mortality associated with acute Plasmodium vivax malaria. Clin Microbiol Rev 26(1). 2013 Jan;36−57. PMID: 23297258.ArticlePubMed
- 5. Anstey N.M., Douglas N.M., Poespoprodjo J.R.. Plasmodium vivax: clinical spectrum, risk factors and pathogenesis. Adv Parasitol 80:2012;151−201. PMID: 23199488.ArticlePubMed
- 6. Genton B., D'Acremont V., Rare L.. Plasmodium vivax and mixed infections are associated with severe malaria in children: a prospective cohort study from Papua New Guinea. PLoS Med 5(6). 2008 Jun;e127PMID: 18563961.ArticlePubMed
- 7. Price R.N., von Seidlein L., Valecha N.. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect Dis 14(10). 2014 Oct;982−991. PMID: 25213732.ArticlePubMed
- 8. Blackman M.J., Heidrich H.G., Donachie S.. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion-inhibiting antibodies. J Exp Med 172(1). 1990 Jul;379−382. PMID: 1694225.ArticlePubMed
- 9. Galinski M.R., Medina C.C., Ingravallo P.. A reticulocyte-binding protein complex of Plasmodium vivax merozoites. Cell 69(7). 1992 Jun;1213−1226. PMID: 1617731.ArticlePubMed
- 10. Holder A.A., Blackman M.J., Burghaus P.A.. A malaria merozoite surface protein (MSP1)-structure, processing and function. Mem Inst Oswaldo Cruz 87(3). 1992 Jan;37−42. PMID: 1343716.Article
- 11. Mourao L.C., Morais C.G., Bueno L.L.. Naturally acquired antibodies to Plasmodium vivax blood-stage vaccine candidates (PvMSP-119 and PvMSP-3α359-798) and their relationship with hematological features in malaria patients from the Brazilian Amazon. Microbes Infect 14(9). 2012 Aug;730−739. PMID: 22445906.ArticlePubMed
- 12. Xia H., Fang Q., Jangpatarapongsa K.. A comparative study of natural immune responses against Plasmodium vivax C-terminal merozoite surface protein-1 (PvMSP-1) and apical membrane antigen-1 (PvAMA-1) in two endemic settings. EXCLI J 14:2015 Dec;926−934. PMID: 26713085.PubMed
- 13. Egan A., Waterfall M., Pinder M.. Characterization of human T- and B-cell epitopes in the C terminus of Plasmodium falciparum merozoite surface protein 1: evidence for poor T-cell recognition of polypeptides with numerous disulfide bonds. Infect Immun 65(8). 1997 Aug;3024−3031. PMID: 9234749.ArticlePubMed
- 14. Han H.J., Park S.G., Kim S.H.. Epidermal growth factor-like motifs 1 and 2 of Plasmodium vivax merozoite surface protein 1 are critical domains in erythrocyte invasion. Biochem Biophys Res Commun 320(2). 2004 Jul;563−570. PMID: 15219866.ArticlePubMed
- 15. Kang J.M., Ju H.L., Kang Y.M.. Genetic polymorphism and natural selection in the C-terminal 42 kDa region of merozoite surface protein-1 among Plasmodium vivax Korean isolates. Malar J 11:2012 Jun;206PMID: 22709605.ArticlePubMed
- 16. del Portillo H.A., Longacre S., Khouri E.. Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc Natl Acad Sci U S A 88(9). 1991 May;4030−4034. PMID: 2023952.ArticlePubMed
- 17. Gibson H.L., Tucker J.E., Kaslow D.C.. Structure and expression of the gene for Pv200, a major blood-stage surface antigen of Plasmodium vivax. Mol Biochem Parasitol 50(2). 1992 Feb;325−333. PMID: 1371329.ArticlePubMed
- 18. Goo Y.K., Moon J.H., Ji S.Y.. The unique distribution of the Plasmodium vivax merozoite surface protein 1 in parasite isolates with short and long latent periods from the Republic of Korea. Malar J 14:2015 Aug;299PMID: 26242878.ArticlePubMed
- 19. Kim J.Y., Suh E.J., Yu H.S.. Longitudinal and cross-sectional genetic diversity in the Korean Peninsula based on the P. vivax merozoite surface protein gene. Osong Public Health Res Perspect 2(3). 2011 Dec;158−163. PMID: 24159467.ArticlePubMed
- 20. Kim J.Y., Goo Y.K., Ji S.Y.. Development and efficacy of real-time PCR in the diagnosis of vivax malaria using field samples in the Republic of Korea. PLoS One 9(8). 2014 Aug;e105871PMID: 25148038.ArticlePubMed
- 21. Snounou G., Viriyakosol S., Zhu X.P.. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol 61(2). 1993 Oct;315−320. PMID: 8264734.ArticlePubMed
- 22. Goo Y.K., Seo E.J., Choi Y.K.. First characterization of Plasmodium vivax liver stage antigen (PvLSA) using synthetic peptides. Parasit Vectors 7:2014 Feb;64PMID: 24520895.ArticlePubMed
- 23. Babon J.J., Morgan W.D., Kelly G.. Structural studies on Plasmodium vivax merozoite surface protein-1. Mol Biochem Parasitol 153(1). 2007 May;31−40. PMID: 17343930.ArticlePubMed
- 24. Mirahmadi H., Fallahi S., Seyyed Tabaei S.J.. Soluble recombinant merozoite surface antigen-142 kDa of Plasmodium vivax: an improved diagnostic antigen for vivax malaria. J Microbiol Methods 123:2016 Apr;44−50. PMID: 26851675.ArticlePubMed
- 25. Soares I.S., Levitus G., Souza J.M.. Acquired immune responses to the N- and C-terminal regions of Plasmodium vivax merozoite surface protein 1 in individuals exposed to malaria. Infect Immun 65(5). 1997 May;1606−1614. PMID: 9125537.ArticlePubMed
- 26. Iwagami M., Fukumoto M., Hwang S.Y.. Population structure and transmission dynamics of Plasmodium vivax in the Republic of Korea based on microsatellite DNA analysis. PLoS Negl Trop Dis 6(4). 2012 Apr;e1592PMID: 22509416.ArticlePubMed
- 27. Iwagami M., Hwang S.Y., Kim S.H.. Microsatellite DNA analysis revealed a drastic genetic change of Plasmodium vivax population in the Republic of Korea during 2002 and 2003. PLoS Negl Trop Dis 7(10). 2013 Nov;e2522PMID: 24205429.ArticlePubMed
- 28. Choi Y.K., Choi K.M., Park M.H.. Rapid dissemination of newly introduced Plasmodium vivax genotypes in South Korea. Am J Trop Med Hyg 82(3). 2010 Mar;426−432. PMID: 20207868.ArticlePubMed
References
Figure 1Diagram and antigenicity evaluation of three recombinant Plasmodium vivax merozoite surface protein 1 (PvMSP1) proteins: rPvMSP1-19, rPvMSP1-33 Sal 1, and rPvMSP1-33 Belem. (A) A diagram of the three recombinant proteins produced. (B) SDS-PAGE of rPvMSP1-19, rPvMSP1-33 Sal 1, and rPvMSP1-33 Belem. Successful expression of these recombinant proteins in Escherichia coli was confirmed using SDS-PAGE and Coomassie blue staining. (C) Western blot analyses of rPvMSP1-19, rPvMSP1-33 Sal 1, and rPvMSP1-33 Belem. Antigenicity and the ability to differentiate specific antibodies against the three recombinant proteins were analyzed via Western blotting with sera obtained from patients infected with PvMSP1-Sal 1 (patient serum-MSP1 Sal 1) and PvMSP1-Belem (patient serum-MSP1 Belem), and sera from healthy individuals (normal serum). Lanes: M, molecular mass marker; 1, rPvMSP1-Belem; 2, rPvMSP1-Sal 1; 3, rPvMSP1-19. SDS-PAGE = sodium dodecyl sulfate-polyacrylamide gel electrophoresis.


Figure 2Levels of antibodies specific to Plasmodium vivax merozoite surface protein 1 (PvMSP1)-19, PvMSP1-23 Sal 1, and PvMSP1-23 Belem among malaria patients in vivax endemic regions of Korea. * p < 0.05. OD = optical density.
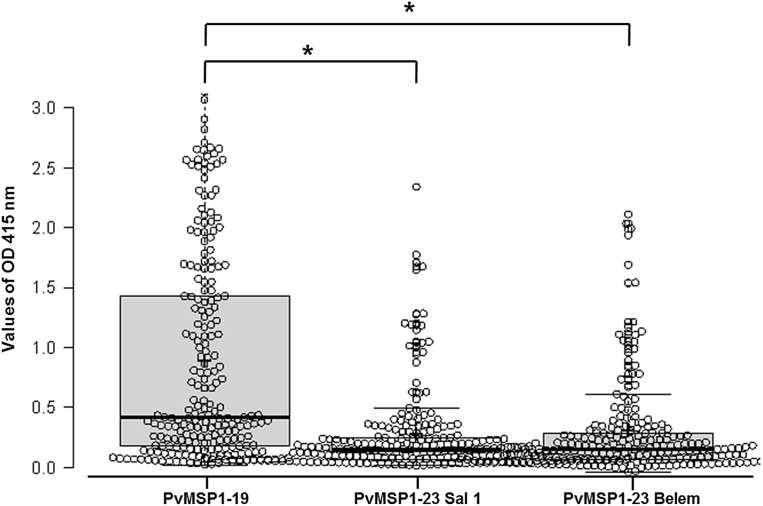

Table 1Seroprevalence of antibodies specific to Plasmodium vivax merozoite surface protein 1 (PvMSP1)-19, PvMSP1-23 Sal 1, and PvMSP1-23 Belem in 221 samples from a Korean population.
| PvMSP1-19* | PvMSP1-23 Sal 1 | PvMSP1-23 Belem | |
|---|---|---|---|
| Positive, n (%) | 198 (89.6) | 142 (64.3) | 106 (48.0) |
| Negative, n (%) | 23 (10.4) | 79 (35.7) | 115 (52.0) |
Table 2Details of the patterns of seropositivity to Plasmodium vivax merozoite surface protein 1 (PvMSP1)-19, PvMSP1-23 Sal 1, and PvMSP1-23 Belem in 221 P. vivax-infected Korean individuals.
Figure & Data
References
Citations
Citations to this article as recorded by 

- Plasmodium vivax MSP1-42 kD Variant Proteins Detected Naturally Induced IgG Antibodies in Patients Regardless of the Infecting Parasite Phenotype in Mesoamerica
Lilia Gonzalez-Ceron, Barbara Dema, Olga L. Palomeque-Culebro, Frida Santillan-Valenzuela, Alberto Montoya, Arturo Reyes-Sandoval
Life.2023; 13(3): 704. CrossRef - Spatiotemporal Changes in Plasmodium vivax msp142 Haplotypes in Southern Mexico: From the Control to the Pre-Elimination Phase
Alejandro Flores-Alanis, Lilia González-Cerón, Frida Santillán-Valenzuela, Cecilia Ximenez, Marco A. Sandoval-Bautista, Rene Cerritos
Microorganisms.2022; 10(1): 186. CrossRef - Diversity and natural selection of Merozoite surface Protein-1 in three species of human malaria parasites: Contribution from South-East Asian isolates
Xiang Ting Goh, Yvonne A.L. Lim, Ping Chin Lee, Veeranoot Nissapatorn, Kek Heng Chua
Molecular and Biochemical Parasitology.2021; 244: 111390. CrossRef - Humoral and cellular immune response to Plasmodium vivax VIR recombinant and synthetic antigens in individuals naturally exposed to P. vivax in the Republic of Korea
Sanghyun Lee, Young-Ki Choi, Youn-Kyoung Goo
Malaria Journal.2021;[Epub] CrossRef


 PubReader
PubReader Cite
Cite
